Structure of the NS3 helicase from Zika virus
- PMID: 27399257
- PMCID: PMC5085289
- DOI: 10.1038/nsmb.3258
Structure of the NS3 helicase from Zika virus
Abstract
Zika virus has emerged as a pathogen of major health concern. Here, we present a high-resolution (1.62-Å) crystal structure of the RNA helicase from the French Polynesia strain. The structure is similar to that of the RNA helicase from Dengue virus, with variability in the conformations of loops typically involved in binding ATP and RNA. We identify druggable 'hotspots' that are well suited for in silico and/or fragment-based high-throughput drug discovery.
Figures
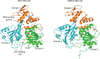
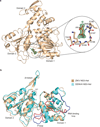
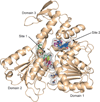
Similar articles
-
The crystal structure of Zika virus helicase: basis for antiviral drug design.Protein Cell. 2016 Jun;7(6):450-4. doi: 10.1007/s13238-016-0275-4. Protein Cell. 2016. PMID: 27172988 Free PMC article. No abstract available.
-
Discovery of Novel Druggable Sites on Zika Virus NS3 Helicase Using X-ray Crystallography-Based Fragment Screening.Int J Mol Sci. 2018 Nov 20;19(11):3664. doi: 10.3390/ijms19113664. Int J Mol Sci. 2018. PMID: 30463319 Free PMC article.
-
Crystal structures of the methyltransferase and helicase from the ZIKA 1947 MR766 Uganda strain.Acta Crystallogr D Struct Biol. 2017 Sep 1;73(Pt 9):767-774. doi: 10.1107/S2059798317010737. Epub 2017 Aug 15. Acta Crystallogr D Struct Biol. 2017. PMID: 28876240
-
Crystal structure of unlinked NS2B-NS3 protease from Zika virus.Science. 2016 Dec 23;354(6319):1597-1600. doi: 10.1126/science.aai9309. Epub 2016 Dec 8. Science. 2016. PMID: 27940580
-
A Perspective on Protein Structure Prediction Using Quantum Computers.J Chem Theory Comput. 2024 May 14;20(9):3359-3378. doi: 10.1021/acs.jctc.4c00067. Epub 2024 May 4. J Chem Theory Comput. 2024. PMID: 38703105 Free PMC article. Review.
Cited by
-
Antiviral activity of lycorine against Zika virus in vivo and in vitro.Virology. 2020 Jul;546:88-97. doi: 10.1016/j.virol.2020.04.009. Epub 2020 Apr 19. Virology. 2020. PMID: 32452420 Free PMC article.
-
Identification of potential inhibitors of Zika virus targeting NS3 helicase using molecular dynamics simulations and DFT studies.Mol Divers. 2023 Aug;27(4):1689-1701. doi: 10.1007/s11030-022-10522-5. Epub 2022 Sep 5. Mol Divers. 2023. PMID: 36063275
-
Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication.Viruses. 2019 Oct 10;11(10):929. doi: 10.3390/v11100929. Viruses. 2019. PMID: 31658680 Free PMC article. Review.
-
Zika virus proteins at an atomic scale: how does structural biology help us to understand and develop vaccines and drugs against Zika virus infection?J Venom Anim Toxins Incl Trop Dis. 2019 Aug 29;25:e20190013. doi: 10.1590/1678-9199-JVATITD-2019-0013. J Venom Anim Toxins Incl Trop Dis. 2019. PMID: 31523227 Free PMC article. Review.
-
Discovery and Computational Analyses of Novel Small Molecule Zika Virus Inhibitors.Molecules. 2019 Apr 13;24(8):1465. doi: 10.3390/molecules24081465. Molecules. 2019. PMID: 31013906 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

