DNA binding triggers tetramerization of the glucocorticoid receptor in live cells
- PMID: 27382178
- PMCID: PMC4961135
- DOI: 10.1073/pnas.1606774113
DNA binding triggers tetramerization of the glucocorticoid receptor in live cells
Abstract
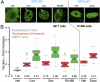
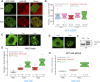
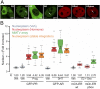
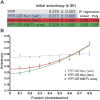
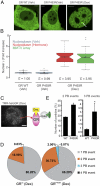
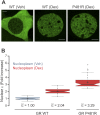
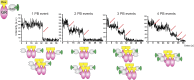
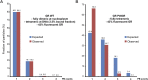
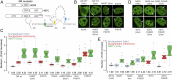
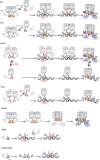
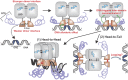
Transcription factors dynamically bind to chromatin and are essential for the regulation of genes. Although a large percentage of these proteins appear to self-associate to form dimers or higher order oligomers, the stoichiometry of DNA-bound transcription factors has been poorly characterized in vivo. The glucocorticoid receptor (GR) is a ligand-regulated transcription factor widely believed to act as a dimer or a monomer. Using a unique set of imaging techniques coupled with a cell line containing an array of DNA binding elements, we show that GR is predominantly a tetramer when bound to its target DNA. We find that DNA binding triggers an interdomain allosteric regulation within the GR, leading to tetramerization. We therefore propose that dynamic changes in GR stoichiometry represent a previously unidentified level of regulation in steroid receptor activation. Quaternary structure analysis of other members of the steroid receptor family (estrogen, androgen, and progesterone receptors) reveals variation in oligomerization states among this family of transcription factors. Because GR's oligomerization state has been implicated in therapy outcome, our findings open new doors to the rational design of novel GR ligands and redefine the quaternary structure of steroid receptors.
Keywords: dimer; glucocorticoid receptor; number and brightness; steroid receptors; tetramer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
More than meets the dimer: What is the quaternary structure of the glucocorticoid receptor?Transcription. 2017 Jan;8(1):32-39. doi: 10.1080/21541264.2016.1249045. Epub 2016 Oct 20. Transcription. 2017. PMID: 27764575 Free PMC article.
-
Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces.Mol Cell Biol. 2001 Feb;21(3):781-93. doi: 10.1128/MCB.21.3.781-793.2001. Mol Cell Biol. 2001. PMID: 11154266 Free PMC article.
-
Glucocorticoid Receptor-DNA Dissociation Kinetics Measured in Vitro Reveal Exchange on the Second Time Scale.Biochemistry. 2015 Sep 1;54(34):5306-14. doi: 10.1021/acs.biochem.5b00693. Epub 2015 Aug 18. Biochemistry. 2015. PMID: 26267475
-
New insights into the anti-inflammatory mechanisms of glucocorticoids: an emerging role for glucocorticoid-receptor-mediated transactivation.Endocrinology. 2013 Mar;154(3):993-1007. doi: 10.1210/en.2012-2045. Epub 2013 Feb 5. Endocrinology. 2013. PMID: 23384835 Review.
-
There goes the neighborhood: Assembly of transcriptional complexes during the regulation of metabolism and inflammation by the glucocorticoid receptor.Steroids. 2016 Oct;114:7-15. doi: 10.1016/j.steroids.2016.05.003. Epub 2016 May 15. Steroids. 2016. PMID: 27192428 Free PMC article. Review.
Cited by
-
Radial pair correlation of molecular brightness fluctuations maps protein diffusion as a function of oligomeric state within live-cell nuclear architecture.Biophys J. 2022 Jun 7;121(11):2152-2167. doi: 10.1016/j.bpj.2022.04.030. Epub 2022 Apr 30. Biophys J. 2022. PMID: 35490296 Free PMC article.
-
Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor.BMC Biol. 2020 Jun 2;18(1):59. doi: 10.1186/s12915-020-00788-2. BMC Biol. 2020. PMID: 32487073 Free PMC article.
-
A General Introduction to Glucocorticoid Biology.Front Immunol. 2019 Jul 4;10:1545. doi: 10.3389/fimmu.2019.01545. eCollection 2019. Front Immunol. 2019. PMID: 31333672 Free PMC article. Review.
-
Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.Nat Struct Mol Biol. 2023 Mar;30(3):286-295. doi: 10.1038/s41594-022-00914-4. Epub 2023 Feb 6. Nat Struct Mol Biol. 2023. PMID: 36747092
-
Roles of the Glucocorticoid and Mineralocorticoid Receptors in Skin Pathophysiology.Int J Mol Sci. 2018 Jun 29;19(7):1906. doi: 10.3390/ijms19071906. Int J Mol Sci. 2018. PMID: 29966221 Free PMC article. Review.
References
-
- Clark AR, Belvisi MG. Maps and legends: The quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther. 2012;134(1):54–67. - PubMed
-
- Ayroldi E, Macchiarulo A, Riccardi C. Targeting glucocorticoid side effects: Selective glucocorticoid receptor modulator or glucocorticoid-induced leucine zipper? A perspective. FASEB J. 2014;28(12):5055–5070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

