All Subdomains of the Talin Rod Are Mechanically Vulnerable and May Contribute To Cellular Mechanosensing
- PMID: 27380548
- PMCID: PMC4982699
- DOI: 10.1021/acsnano.6b01658
All Subdomains of the Talin Rod Are Mechanically Vulnerable and May Contribute To Cellular Mechanosensing
Abstract
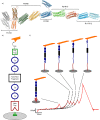
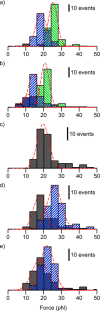
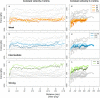
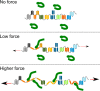
Although the relevance of mechanotransduction in cell signaling is currently appreciated, the mechanisms that drive this process remain largely unknown. Mechanical unfolding of proteins may trigger distinct downstream signals in cells, providing a mechanism for cellular mechanotransduction. Force-induced unfolding of talin, a prominent focal adhesion protein, has been demonstrated previously for a small portion of its rod domain. Here, using single-molecule atomic force microscopy (smAFM), we show that the entire talin rod can be unfolded by mechanical extension, over a physiological range of forces between 10 and 40 pN. We also demonstrate, through a combination of smAFM and steered molecular dynamics, that the different bundles within the talin rod exhibit a distinct hierarchy of mechanical stability. These results provide a mechanism by which different force conditions within the cell control a graduated unfolding of the talin rod. Mechanical unfolding of the rod subdomains, and the subsequent effect on talin's binding interactions, would allow for a finely tuned cellular response to internally or externally applied forces.
Keywords: mechanobiology; mechanotransduction; protein mechanostability; single-molecule force spectroscopy; steered molecular dynamics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Talin-mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling.J Cell Sci. 2019 Apr 3;132(7):jcs226514. doi: 10.1242/jcs.226514. J Cell Sci. 2019. PMID: 30837291
-
Mechanical unfolding reveals stable 3-helix intermediates in talin and α-catenin.PLoS Comput Biol. 2018 Apr 26;14(4):e1006126. doi: 10.1371/journal.pcbi.1006126. eCollection 2018 Apr. PLoS Comput Biol. 2018. PMID: 29698481 Free PMC article.
-
Mechanical Unfolding of Proteins-A Comparative Nonequilibrium Molecular Dynamics Study.Biophys J. 2020 Sep 1;119(5):939-949. doi: 10.1016/j.bpj.2020.07.030. Epub 2020 Aug 6. Biophys J. 2020. PMID: 32822586 Free PMC article.
-
Talin Dependent Mechanosensitivity of Cell Focal Adhesions.Cell Mol Bioeng. 2015;8(1):151-159. doi: 10.1007/s12195-014-0364-5. Epub 2014 Nov 4. Cell Mol Bioeng. 2015. PMID: 26097520 Free PMC article. Review.
-
Talin‑1 interaction network in cellular mechanotransduction (Review).Int J Mol Med. 2022 May;49(5):60. doi: 10.3892/ijmm.2022.5116. Epub 2022 Mar 10. Int J Mol Med. 2022. PMID: 35266014 Free PMC article. Review.
Cited by
-
Molecular dynamics simulations reveal how vinculin refolds partially unfolded talin rod helices to stabilize them against mechanical force.PLoS Comput Biol. 2024 Aug 7;20(8):e1012341. doi: 10.1371/journal.pcbi.1012341. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39110765 Free PMC article.
-
The mechanical cell - the role of force dependencies in synchronising protein interaction networks.J Cell Sci. 2022 Nov 15;135(22):jcs259769. doi: 10.1242/jcs.259769. Epub 2022 Nov 18. J Cell Sci. 2022. PMID: 36398718 Free PMC article. Review.
-
A HaloTag-TEV genetic cassette for mechanical phenotyping of proteins from tissues.Nat Commun. 2020 Apr 28;11(1):2060. doi: 10.1038/s41467-020-15465-9. Nat Commun. 2020. PMID: 32345978 Free PMC article.
-
Talin: a protein designed for mechanotransduction.Emerg Top Life Sci. 2018 Dec 21;2(5):673-675. doi: 10.1042/ETLS20180179. Emerg Top Life Sci. 2018. PMID: 33530661 Free PMC article.
-
Molecular Fluctuations as a Ruler of Force-Induced Protein Conformations.Nano Lett. 2021 Apr 14;21(7):2953-2961. doi: 10.1021/acs.nanolett.1c00051. Epub 2021 Mar 25. Nano Lett. 2021. PMID: 33765390 Free PMC article.
References
-
- Balaban N. Q.; Schwarz U. S.; Riveline D.; Goichberg P.; Tzur G.; Sabanay I.; Mahalu D.; Safran S.; Bershadsky A.; Addadi L.; Geiger B. Force and Focal Adhesion Assembly: A Close Relationship Studied Using Elastic Micropatterned Substrates. Nat. Cell Biol. 2001, 3, 466–472. 10.1038/35074532. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

