Molecular Characterization and Transcriptional Regulation Analysis of the Bovine PDHB Gene
- PMID: 27379520
- PMCID: PMC4933360
- DOI: 10.1371/journal.pone.0157445
Molecular Characterization and Transcriptional Regulation Analysis of the Bovine PDHB Gene
Abstract
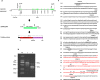
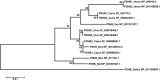
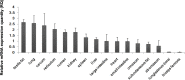
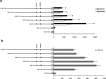
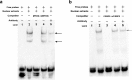
The pyruvate dehydrogenase beta subunit (PDHB) is a subunit of pyruvate dehydrogenase (E1), which catalyzes pyruvate into acetyl-CoA and provides a linkage between the tricarboxylic acid cycle (TCA) and the glycolysis pathway. Previous studies demonstrated PDHB to be positively related to the intramuscular fat (IMF) content. However, the transcriptional regulation of PDHB remains unclear. In our present study, the cDNA of bovine PDHB was cloned and the genomic structure was analyzed. The phylogenetic tree showed bovine PDHB to be closely related to goat and sheep, and least related to chicken. Spatial expression pattern analysis revealed the products of bovine PDHB to be widely expressed with the highest level in the fat of testis. To understand the transcriptional regulation of bovine PDHB, 1899 base pairs (bp) of the 5'-regulatory region was cloned. Sequence analysis neither found consensus TATA-box nor CCAAT-box in the 5'-flanking region of bovine PDHB. However, a CpG island was predicted from nucleotides -284 to +117. Serial deletion constructs of the 5'-flanking region, evaluated in dual-luciferase reporter assay, revealed the core promoter to be located 490bp upstream from the transcription initiation site (+1). Electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation assay (ChIP) in combination with asite-directed mutation experiment indicated both myogenin (MYOG) and the CCAAT/enhancer-binding protein beta (C/EBPß) to be important transcription factors for bovine PDHB in skeletal muscle cells and adipocytes. Our results provide an important basis for further investigation of the bovine PDHB function and regulation in cattle.
Conflict of interest statement
Figures



Similar articles
-
Tissue expression analysis, cloning and characterization of the 5'-regulatory region of the bovine FABP3 gene.Mol Biol Rep. 2016 Sep;43(9):991-8. doi: 10.1007/s11033-016-4026-7. Epub 2016 Jun 6. Mol Biol Rep. 2016. PMID: 27270359
-
Dual promoter structure of ZFP106: regulation by myogenin and nuclear respiratory factor-1.Gene. 2005 Jan 3;344:143-59. doi: 10.1016/j.gene.2004.09.035. Epub 2004 Nov 19. Gene. 2005. PMID: 15656981
-
Characterization of the infection-responsive bovine lactoferrin promoter.Gene. 2005 Jun 20;353(1):107-17. doi: 10.1016/j.gene.2005.04.016. Gene. 2005. PMID: 15935571
-
Transcriptional regulation of the mouse PNRC2 promoter by the nuclear factor Y (NFY) and E2F1.Gene. 2005 Nov 21;361:89-100. doi: 10.1016/j.gene.2005.07.012. Epub 2005 Sep 21. Gene. 2005. PMID: 16181749
-
Characterization of the rat RALDH1 promoter. A functional CCAAT and octamer motif are critical for basal promoter activity.Biochim Biophys Acta. 2002 Dec 12;1579(2-3):81-91. doi: 10.1016/s0167-4781(02)00510-9. Biochim Biophys Acta. 2002. PMID: 12427543
Cited by
-
The Molecular Characteristics of the FAM13A Gene and the Role of Transcription Factors ACSL1 and ASCL2 in Its Core Promoter Region.Genes (Basel). 2019 Nov 28;10(12):981. doi: 10.3390/genes10120981. Genes (Basel). 2019. PMID: 31795267 Free PMC article.
-
Cuproptosis and cuproptosis-related genes in rheumatoid arthritis: Implication, prospects, and perspectives.Front Immunol. 2022 Aug 4;13:930278. doi: 10.3389/fimmu.2022.930278. eCollection 2022. Front Immunol. 2022. PMID: 35990673 Free PMC article. Review.
-
NRF1 and ZSCAN10 bind to the promoter region of the SIX1 gene and their effects body measurements in Qinchuan cattle.Sci Rep. 2017 Aug 11;7(1):7867. doi: 10.1038/s41598-017-08384-1. Sci Rep. 2017. PMID: 28801681 Free PMC article.
-
Unravelling diagnostic clusters and immune landscapes of cuproptosis patterns in intervertebral disc degeneration through dry and wet experiments.Aging (Albany NY). 2023 Dec 29;15(24):15599-15623. doi: 10.18632/aging.205449. Epub 2023 Dec 29. Aging (Albany NY). 2023. PMID: 38159257 Free PMC article.
-
Pyruvate dehydrogenase B regulates myogenic differentiation via the FoxP1-Arih2 axis.J Cachexia Sarcopenia Muscle. 2023 Feb;14(1):606-621. doi: 10.1002/jcsm.13166. Epub 2022 Dec 23. J Cachexia Sarcopenia Muscle. 2023. PMID: 36564038 Free PMC article.
References
-
- Patel MS, Korotchkina LG Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006; 34: 217–222. - PubMed
-
- Mercimek-Mahmutoglu S, Patel J, Cordeiro D, Hewson S, Callen D, Donner EJ, et al. Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia. 2015. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

