EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor
- PMID: 27356095
- PMCID: PMC5027137
- DOI: 10.1021/acschembio.6b00182
EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor
Abstract
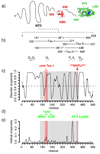
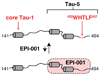
Castration-resistant prostate cancer is the lethal condition suffered by prostate cancer patients that become refractory to androgen deprivation therapy. EPI-001 is a recently identified compound active against this condition that modulates the activity of the androgen receptor, a nuclear receptor that is essential for disease progression. The mechanism by which this compound exerts its inhibitory activity is however not yet fully understood. Here we show, by using high resolution solution nuclear magnetic resonance spectroscopy, that EPI-001 selectively interacts with a partially folded region of the transactivation domain of the androgen receptor, known as transactivation unit 5, that is key for the ability of prostate cells to proliferate in the absence of androgens, a distinctive feature of castration-resistant prostate cancer. Our results can contribute to the development of more potent and less toxic novel androgen receptor antagonists for treating this disease.
Figures

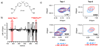
Similar articles
-
Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor.Cancer Cell. 2010 Jun 15;17(6):535-46. doi: 10.1016/j.ccr.2010.04.027. Cancer Cell. 2010. PMID: 20541699
-
An androgen receptor N-terminal domain antagonist for treating prostate cancer.J Clin Invest. 2013 Jul;123(7):2948-60. doi: 10.1172/JCI66398. Epub 2013 Jun 3. J Clin Invest. 2013. PMID: 23722902 Free PMC article.
-
EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer.Oncotarget. 2015 Feb 28;6(6):3811-24. doi: 10.18632/oncotarget.2924. Oncotarget. 2015. PMID: 25669987 Free PMC article.
-
Role of androgen receptor splice variants, their clinical relevance and treatment options.World J Urol. 2020 Mar;38(3):647-656. doi: 10.1007/s00345-018-02619-0. Epub 2019 Jan 19. World J Urol. 2020. PMID: 30659302 Review.
-
Androgen action in the prostate gland.Minerva Urol Nefrol. 2012 Mar;64(1):35-49. Minerva Urol Nefrol. 2012. PMID: 22402316 Review.
Cited by
-
Modulating biomolecular condensates: a novel approach to drug discovery.Nat Rev Drug Discov. 2022 Nov;21(11):841-862. doi: 10.1038/s41573-022-00505-4. Epub 2022 Aug 16. Nat Rev Drug Discov. 2022. PMID: 35974095 Free PMC article. Review.
-
α-Mangostin Promotes In Vitro and In Vivo Degradation of Androgen Receptor and AR-V7 Splice Variant in Prostate Cancer Cells.Cancers (Basel). 2023 Apr 1;15(7):2118. doi: 10.3390/cancers15072118. Cancers (Basel). 2023. PMID: 37046780 Free PMC article.
-
Androgen receptor nucleocytoplasmic trafficking - A one-way journey.Mol Cell Endocrinol. 2023 Oct 1;576:112009. doi: 10.1016/j.mce.2023.112009. Epub 2023 Jul 4. Mol Cell Endocrinol. 2023. PMID: 37414131 Free PMC article. Review.
-
An Overview of Next-Generation Androgen Receptor-Targeted Therapeutics in Development for the Treatment of Prostate Cancer.Int J Mol Sci. 2021 Feb 20;22(4):2124. doi: 10.3390/ijms22042124. Int J Mol Sci. 2021. PMID: 33672769 Free PMC article. Review.
-
Synthesis and Evaluation of Small Molecule Inhibitors of the Androgen Receptor N-Terminal Domain.ACS Med Chem Lett. 2023 Nov 17;14(12):1800-1806. doi: 10.1021/acsmedchemlett.3c00426. eCollection 2023 Dec 14. ACS Med Chem Lett. 2023. PMID: 38116409 Free PMC article.
References
-
- Gelmann EP. Molecular Biology of the Androgen Receptor. J Clin Oncol. 2002;20:3001–3015. - PubMed
-
- Akaza H, Hinotsu S, Usami M, Arai Y, Kanetake H, Naito S, Hirao Y. Combined androgen blockade with bicalutamide for advanced prostate cancer. Cancer. 2009;115:3437–3445. - PubMed
-
- Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002;277:20079–20086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

