De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data
- PMID: 27345837
- PMCID: PMC4985539
- DOI: 10.1016/j.stem.2016.05.010
De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data
Abstract
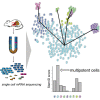
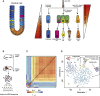
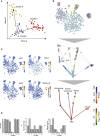
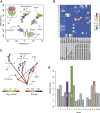
Adult mitotic tissues like the intestine, skin, and blood undergo constant turnover throughout the life of an organism. Knowing the identity of the stem cell is crucial to understanding tissue homeostasis and its aberrations upon disease. Here we present a computational method for the derivation of a lineage tree from single-cell transcriptome data. By exploiting the tree topology and the transcriptome composition, we establish StemID, an algorithm for identifying stem cells among all detectable cell types within a population. We demonstrate that StemID recovers two known adult stem cell populations, Lgr5+ cells in the small intestine and hematopoietic stem cells in the bone marrow. We apply StemID to predict candidate multipotent cell populations in the human pancreas, a tissue with largely uncharacterized turnover dynamics. We hope that StemID will accelerate the search for novel stem cells by providing concrete markers for biological follow-up and validation.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Clonal interrogation of stem cells.Nat Methods. 2011 Apr;8(4 Suppl):S36-40. doi: 10.1038/nmeth.1590. Nat Methods. 2011. PMID: 21451511 Review.
-
Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell.Nature. 2019 May;569(7754):121-125. doi: 10.1038/s41586-019-1154-y. Epub 2019 Apr 24. Nature. 2019. PMID: 31019301
-
Isolation and assessment of long-term reconstituting hematopoietic stem cells from adult mouse bone marrow.Curr Protoc Stem Cell Biol. 2007 Nov;Chapter 2:Unit 2A.4. doi: 10.1002/9780470151808.sc02a04s3. Curr Protoc Stem Cell Biol. 2007. PMID: 18785176
-
A method for isolating pluripotent/multipotent stem cells from blood by using the pluripotent and germ-line DAZL gene as a marker.Stem Cells Dev. 2009 Nov;18(9):1263-71. doi: 10.1089/scd.2008.0406. Stem Cells Dev. 2009. PMID: 19327014
-
Heterogeneous populations of bone marrow stem cells--are we spotting on the same cells from the different angles?Folia Histochem Cytobiol. 2004;42(3):139-46. Folia Histochem Cytobiol. 2004. PMID: 15493574 Review.
Cited by
-
Human genetics uncovers MAP3K15 as an obesity-independent therapeutic target for diabetes.Sci Adv. 2022 Nov 18;8(46):eadd5430. doi: 10.1126/sciadv.add5430. Epub 2022 Nov 16. Sci Adv. 2022. PMID: 36383675 Free PMC article.
-
HumanIslets: An integrated platform for human islet data access and analysis.bioRxiv [Preprint]. 2024 Jun 22:2024.06.19.599613. doi: 10.1101/2024.06.19.599613. bioRxiv. 2024. PMID: 38948734 Free PMC article. Preprint.
-
De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution.Nat Commun. 2022 Oct 30;13(1):6498. doi: 10.1038/s41467-022-34271-z. Nat Commun. 2022. PMID: 36310179 Free PMC article.
-
Single cell RNA-seq reveals profound transcriptional similarity between Barrett's oesophagus and oesophageal submucosal glands.Nat Commun. 2018 Oct 15;9(1):4261. doi: 10.1038/s41467-018-06796-9. Nat Commun. 2018. PMID: 30323168 Free PMC article.
-
Single-cell analyses reveal early thymic progenitors and pre-B cells in zebrafish.J Exp Med. 2022 Sep 5;219(9):e20220038. doi: 10.1084/jem.20220038. Epub 2022 Aug 8. J Exp Med. 2022. PMID: 35938989 Free PMC article.
References
-
- Anavy L., Levin M., Khair S., Nakanishi N., Fernandez-Valverde S.L., Degnan B.M., Yanai I. BLIND ordering of large-scale transcriptomic developmental timecourses. Development. 2014;141:1161–1166. - PubMed
-
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. - PubMed
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

