Rhizobial gibberellin negatively regulates host nodule number
- PMID: 27307029
- PMCID: PMC4910070
- DOI: 10.1038/srep27998
Rhizobial gibberellin negatively regulates host nodule number
Abstract
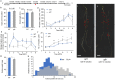
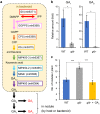
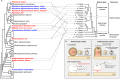
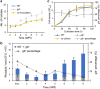
In legume-rhizobia symbiosis, the nodule number is controlled to ensure optimal growth of the host. In Lotus japonicus, the nodule number has been considered to be tightly regulated by host-derived phytohormones and glycopeptides. However, we have discovered a symbiont-derived phytohormonal regulation of nodule number in Mesorhizobium loti. In this study, we found that M. loti synthesized gibberellic acid (GA) under symbiosis. Hosts inoculated with a GA-synthesis-deficient M. loti mutant formed more nodules than those inoculated with the wild-type form at four weeks post inoculation, indicating that GA from already-incorporated rhizobia prevents new nodule formation. Interestingly, the genes for GA synthesis are only found in rhizobial species that inhabit determinate nodules. Our findings suggest that the already-incorporated rhizobia perform GA-associated negative regulation of nodule number to prevent delayed infection by other rhizobia.
Figures

Similar articles
-
Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in Lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific Mesorhizobium loti strains.Plant J. 2018 Jan;93(1):5-16. doi: 10.1111/tpj.13759. Epub 2017 Dec 2. Plant J. 2018. PMID: 29086445
-
Nitric Oxide Detoxification by Mesorhizobium loti Affects Root Nodule Symbiosis with Lotus japonicus.Microbes Environ. 2021;36(3):ME21038. doi: 10.1264/jsme2.ME21038. Microbes Environ. 2021. PMID: 34470944 Free PMC article.
-
Endogenous gibberellins affect root nodule symbiosis via transcriptional regulation of NODULE INCEPTION in Lotus japonicus.Plant J. 2021 Mar;105(6):1507-1520. doi: 10.1111/tpj.15128. Epub 2021 Jan 9. Plant J. 2021. PMID: 33300204
-
Hormone modulation of legume-rhizobial symbiosis.J Integr Plant Biol. 2018 Aug;60(8):632-648. doi: 10.1111/jipb.12653. Epub 2018 Jun 5. J Integr Plant Biol. 2018. PMID: 29578639 Review.
-
Mechanisms in plant growth-promoting rhizobacteria that enhance legume-rhizobial symbioses.J Appl Microbiol. 2020 Nov;129(5):1133-1156. doi: 10.1111/jam.14754. Epub 2020 Jul 24. J Appl Microbiol. 2020. PMID: 32592603 Review.
Cited by
-
The Current Status of Research on Gibberellin Biosynthesis.Plant Cell Physiol. 2020 Dec 23;61(11):1832-1849. doi: 10.1093/pcp/pcaa092. Plant Cell Physiol. 2020. PMID: 32652020 Free PMC article. Review.
-
Unraveling a Tangled Skein: Evolutionary Analysis of the Bacterial Gibberellin Biosynthetic Operon.mSphere. 2020 Jun 3;5(3):e00292-20. doi: 10.1128/mSphere.00292-20. mSphere. 2020. PMID: 32493722 Free PMC article.
-
Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation.J Exp Bot. 2018 Apr 9;69(8):2117-2130. doi: 10.1093/jxb/ery046. J Exp Bot. 2018. PMID: 29432555 Free PMC article.
-
Symbiosis islands of Loteae-nodulating Mesorhizobium comprise three radiating lineages with concordant nod gene complements and nodulation host-range groupings.Microb Genom. 2020 Sep;6(9):mgen000426. doi: 10.1099/mgen.0.000426. Epub 2020 Aug 26. Microb Genom. 2020. PMID: 32845829 Free PMC article.
-
Production of the plant hormone gibberellin by rhizobia increases host legume nodule size.ISME J. 2022 Jul;16(7):1809-1817. doi: 10.1038/s41396-022-01236-5. Epub 2022 Apr 12. ISME J. 2022. PMID: 35414717 Free PMC article.
References
-
- Ferguson B. J. & Mathesius U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 40, 770–790 (2014). - PubMed
-
- Caetano-Anolles G. & Gresshoff P. M. Plant genetic control of nodulation. Annu. Rev. Microbiol. 45, 345–382 (1991). - PubMed
-
- Oka-Kira E. & Kawaguchi M. Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9, 496–502 (2006). - PubMed
-
- Ferguson B. J. et al.. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

