Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells
- PMID: 27300435
- PMCID: PMC5968455
- DOI: 10.1016/j.ccell.2016.05.002
Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells
Abstract
Glioblastomas (GBM) grow in a rich neurochemical milieu, but the impact of neurochemicals on GBM growth is largely unexplored. We interrogated 680 neurochemical compounds in patient-derived GBM neural stem cells (GNS) to determine the effects on proliferation and survival. Compounds that modulate dopaminergic, serotonergic, and cholinergic signaling pathways selectively affected GNS growth. In particular, dopamine receptor D4 (DRD4) antagonists selectively inhibited GNS growth and promoted differentiation of normal neural stem cells. DRD4 antagonists inhibited the downstream effectors PDGFRβ, ERK1/2, and mTOR and disrupted the autophagy-lysosomal pathway, leading to accumulation of autophagic vacuoles followed by G0/G1 arrest and apoptosis. These results demonstrate a role for neurochemical pathways in governing GBM stem cell proliferation and suggest therapeutic approaches for GBM.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
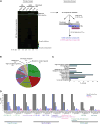
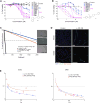
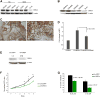
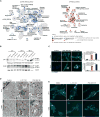
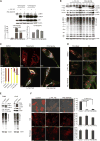

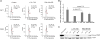

Comment in
-
Gliomas "Dope Up" for Growth.Cancer Cell. 2016 Jun 13;29(6):778-780. doi: 10.1016/j.ccell.2016.05.010. Cancer Cell. 2016. PMID: 27300432
Similar articles
-
Phostine PST3.1a Targets MGAT5 and Inhibits Glioblastoma-Initiating Cell Invasiveness and Proliferation.Mol Cancer Res. 2017 Oct;15(10):1376-1387. doi: 10.1158/1541-7786.MCR-17-0120. Epub 2017 Jun 20. Mol Cancer Res. 2017. PMID: 28634226
-
A high-content small molecule screen identifies sensitivity of glioblastoma stem cells to inhibition of polo-like kinase 1.PLoS One. 2013 Oct 30;8(10):e77053. doi: 10.1371/journal.pone.0077053. eCollection 2013. PLoS One. 2013. PMID: 24204733 Free PMC article.
-
Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells.Oncotarget. 2015 Sep 22;6(28):26192-215. doi: 10.18632/oncotarget.4613. Oncotarget. 2015. PMID: 26295306 Free PMC article.
-
The Multi-Faceted Effect of Curcumin in Glioblastoma from Rescuing Cell Clearance to Autophagy-Independent Effects.Molecules. 2020 Oct 20;25(20):4839. doi: 10.3390/molecules25204839. Molecules. 2020. PMID: 33092261 Free PMC article. Review.
-
Progress on potential strategies to target brain tumor stem cells.Cell Mol Neurobiol. 2009 Mar;29(2):141-55. doi: 10.1007/s10571-008-9310-1. Epub 2008 Sep 10. Cell Mol Neurobiol. 2009. PMID: 18781384 Review.
Cited by
-
Identification of a ferroptosis-related gene signature predictive model in colon cancer.World J Surg Oncol. 2021 Apr 29;19(1):135. doi: 10.1186/s12957-021-02244-z. World J Surg Oncol. 2021. PMID: 33926457 Free PMC article.
-
A Novel Multi-Target Small Molecule, LCC-09, Inhibits Stemness and Therapy-Resistant Phenotypes of Glioblastoma Cells by Increasing miR-34a and Deregulating the DRD4/Akt/mTOR Signaling Axis.Cancers (Basel). 2019 Sep 26;11(10):1442. doi: 10.3390/cancers11101442. Cancers (Basel). 2019. PMID: 31561595 Free PMC article.
-
Overexpression of Parkin in the Neuronal Progenitor Cells from a Patient with Parkinson's Disease Shifts the Transcriptome Towards the Normal State.Mol Neurobiol. 2023 Jun;60(6):3522-3533. doi: 10.1007/s12035-023-03293-z. Epub 2023 Mar 8. Mol Neurobiol. 2023. PMID: 36884134
-
Therapeutic advantage of targeting lysosomal membrane integrity supported by lysophagy in malignant glioma.Cancer Sci. 2022 Aug;113(8):2716-2726. doi: 10.1111/cas.15451. Epub 2022 Jun 27. Cancer Sci. 2022. PMID: 35657693 Free PMC article.
-
The Prospective Value of Dopamine Receptors on Bio-Behavior of Tumor.J Cancer. 2019 Mar 3;10(7):1622-1632. doi: 10.7150/jca.27780. eCollection 2019. J Cancer. 2019. PMID: 31205518 Free PMC article. Review.
References
-
- Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Bryja V, Halliez S, Nishimaru H, et al. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. - PubMed
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radio-resistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
-
- Borta A, Hoglinger GU. Dopamine and adult neurogenesis. J. Neurochem. 2007;100:587–595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

