Probing the impact of chromatin conformation on genome editing tools
- PMID: 27280977
- PMCID: PMC5291272
- DOI: 10.1093/nar/gkw524
Probing the impact of chromatin conformation on genome editing tools
Abstract
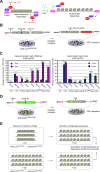
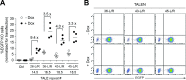
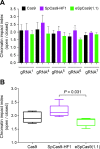
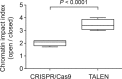
Transcription activator-like effector nucleases (TALENs) and RNA-guided nucleases derived from clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 systems have become ubiquitous genome editing tools. Despite this, the impact that distinct high-order chromatin conformations have on these sequence-specific designer nucleases is, presently, ill-defined. The same applies to the relative performance of TALENs and CRISPR/Cas9 nucleases at isogenic target sequences subjected to different epigenetic modifications. Here, to address these gaps in our knowledge, we have implemented quantitative cellular systems based on genetic reporters in which the euchromatic and heterochromatic statuses of designer nuclease target sites are stringently controlled by small-molecule drug availability. By using these systems, we demonstrate that TALENs and CRISPR/Cas9 nucleases are both significantly affected by the high-order epigenetic context of their target sequences. In addition, this outcome could also be ascertained for S. pyogenes CRISPR/Cas9 complexes harbouring Cas9 variants whose DNA cleaving specificities are superior to that of the wild-type Cas9 protein. Thus, the herein investigated cellular models will serve as valuable functional readouts for screening and assessing the role of chromatin on designer nucleases based on different platforms or with different architectures or compositions.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications.Mol Plant. 2016 Jul 6;9(7):961-74. doi: 10.1016/j.molp.2016.04.009. Epub 2016 Apr 20. Mol Plant. 2016. PMID: 27108381 Review.
-
Applications of Alternative Nucleases in the Age of CRISPR/Cas9.Int J Mol Sci. 2017 Nov 29;18(12):2565. doi: 10.3390/ijms18122565. Int J Mol Sci. 2017. PMID: 29186020 Free PMC article. Review.
-
Genome editing: the road of CRISPR/Cas9 from bench to clinic.Exp Mol Med. 2016 Oct 14;48(10):e265. doi: 10.1038/emm.2016.111. Exp Mol Med. 2016. PMID: 27741224 Free PMC article. Review.
-
CRISPR/Cas9: an advanced tool for editing plant genomes.Transgenic Res. 2016 Oct;25(5):561-73. doi: 10.1007/s11248-016-9953-5. Epub 2016 Mar 24. Transgenic Res. 2016. PMID: 27012546 Review.
-
Basics of genome editing technology and its application in livestock species.Reprod Domest Anim. 2017 Aug;52 Suppl 3:4-13. doi: 10.1111/rda.13012. Reprod Domest Anim. 2017. PMID: 28815851 Review.
Cited by
-
Decorating chromatin for enhanced genome editing using CRISPR-Cas9.Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2204259119. doi: 10.1073/pnas.2204259119. Epub 2022 Dec 2. Proc Natl Acad Sci U S A. 2022. PMID: 36459645 Free PMC article.
-
Gene Editing Technologies to Target HBV cccDNA.Viruses. 2022 Nov 28;14(12):2654. doi: 10.3390/v14122654. Viruses. 2022. PMID: 36560658 Free PMC article. Review.
-
Improving the on-target activity of high-fidelity Cas9 editors by combining rational design and random mutagenesis.Appl Microbiol Biotechnol. 2023 Apr;107(7-8):2385-2401. doi: 10.1007/s00253-023-12469-5. Epub 2023 Mar 14. Appl Microbiol Biotechnol. 2023. PMID: 36917274
-
Inhibition of complement activation by CD55 overexpression in human induced pluripotent stem cell derived kidney organoids.Front Immunol. 2023 Jan 12;13:1058763. doi: 10.3389/fimmu.2022.1058763. eCollection 2022. Front Immunol. 2023. PMID: 36713440 Free PMC article.
-
Epigenetic features improve TALE target prediction.BMC Genomics. 2021 Dec 29;22(1):914. doi: 10.1186/s12864-021-08210-z. BMC Genomics. 2021. PMID: 34965853 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

