Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition
- PMID: 27239761
- PMCID: PMC5916782
- DOI: 10.1038/nm.4105
Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition
Erratum in
-
Corrigendum: Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition.Nat Med. 2016 Oct 6;22(10):1192. doi: 10.1038/nm1016-1192a. Nat Med. 2016. PMID: 27711066 No abstract available.
Abstract
A recombinant vaccine containing Aventis Pasteur's canarypox vector (ALVAC)-HIV and gp120 alum decreased the risk of HIV acquisition in the RV144 vaccine trial. The substitution of alum with the more immunogenic MF59 adjuvant is under consideration for the next efficacy human trial. We found here that an ALVAC-simian immunodeficiency virus (SIV) and gp120 alum (ALVAC-SIV + gp120) equivalent vaccine, but not an ALVAC-SIV + gp120 MF59 vaccine, was efficacious in delaying the onset of SIVmac251 in rhesus macaques, despite the higher immunogenicity of the latter adjuvant. Vaccine efficacy was associated with alum-induced, but not with MF59-induced, envelope (Env)-dependent mucosal innate lymphoid cells (ILCs) that produce interleukin (IL)-17, as well as with mucosal IgG to the gp120 variable region 2 (V2) and the expression of 12 genes, ten of which are part of the RAS pathway. The association between RAS activation and vaccine efficacy was also observed in an independent efficacious SIV-vaccine approach. Whether RAS activation, mucosal ILCs and antibodies to V2 are also important hallmarks of HIV-vaccine efficacy in humans will require further studies.
Figures
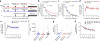


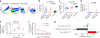


Similar articles
-
Expression of CD40L by the ALVAC-Simian Immunodeficiency Virus Vector Abrogates T Cell Responses in Macaques.J Virol. 2020 Feb 28;94(6):e01933-19. doi: 10.1128/JVI.01933-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896599 Free PMC article.
-
Analysis of Complement-Mediated Lysis of Simian Immunodeficiency Virus (SIV) and SIV-Infected Cells Reveals Sex Differences in Vaccine-Induced Immune Responses in Rhesus Macaques.J Virol. 2018 Sep 12;92(19):e00721-18. doi: 10.1128/JVI.00721-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021899 Free PMC article.
-
Adjuvanting a Simian Immunodeficiency Virus Vaccine with Toll-Like Receptor Ligands Encapsulated in Nanoparticles Induces Persistent Antibody Responses and Enhanced Protection in TRIM5α Restrictive Macaques.J Virol. 2017 Jan 31;91(4):e01844-16. doi: 10.1128/JVI.01844-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928002 Free PMC article.
-
Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIVmac251 Rectal Challenge.PLoS Pathog. 2015 Aug 12;11(8):e1005101. doi: 10.1371/journal.ppat.1005101. eCollection 2015 Aug. PLoS Pathog. 2015. PMID: 26267144 Free PMC article.
-
The role of immunity in protection from mucosal SIV infection in macaques.Oral Dis. 2002;8 Suppl 2:63-8. doi: 10.1034/j.1601-0825.2002.00014.x. Oral Dis. 2002. PMID: 12164663 Review.
Cited by
-
Rapid Induction of Multifunctional Antibodies in Rabbits and Macaques by Clade C HIV-1 CAP257 Envelopes Circulating During Epitope-Specific Neutralization Breadth Development.Front Immunol. 2020 Jun 2;11:984. doi: 10.3389/fimmu.2020.00984. eCollection 2020. Front Immunol. 2020. PMID: 32582155 Free PMC article.
-
Differential Effect of Mucosal NKp44+ Innate Lymphoid Cells and Δγ Cells on Simian Immunodeficiency Virus Infection Outcome in Rhesus Macaques.J Immunol. 2019 Nov 1;203(9):2459-2471. doi: 10.4049/jimmunol.1900572. Epub 2019 Sep 25. J Immunol. 2019. PMID: 31554692 Free PMC article.
-
Co-immunization of DNA and Protein in the Same Anatomical Sites Induces Superior Protective Immune Responses against SHIV Challenge.Cell Rep. 2020 May 12;31(6):107624. doi: 10.1016/j.celrep.2020.107624. Cell Rep. 2020. PMID: 32402293 Free PMC article.
-
Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV.Curr Opin HIV AIDS. 2019 Jul;14(4):309-317. doi: 10.1097/COH.0000000000000551. Curr Opin HIV AIDS. 2019. PMID: 30994501 Free PMC article. Review.
-
Control of Heterologous Simian Immunodeficiency Virus SIVsmE660 Infection by DNA and Protein Coimmunization Regimens Combined with Different Toll-Like-Receptor-4-Based Adjuvants in Macaques.J Virol. 2018 Jul 17;92(15):e00281-18. doi: 10.1128/JVI.00281-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793957 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

