Mechanisms of Antibiotic Resistance
- PMID: 27227291
- PMCID: PMC4888801
- DOI: 10.1128/microbiolspec.VMBF-0016-2015
Mechanisms of Antibiotic Resistance
Abstract
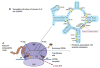
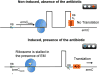
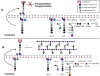
Emergence of resistance among the most important bacterial pathogens is recognized as a major public health threat affecting humans worldwide. Multidrug-resistant organisms have not only emerged in the hospital environment but are now often identified in community settings, suggesting that reservoirs of antibiotic-resistant bacteria are present outside the hospital. The bacterial response to the antibiotic "attack" is the prime example of bacterial adaptation and the pinnacle of evolution. "Survival of the fittest" is a consequence of an immense genetic plasticity of bacterial pathogens that trigger specific responses that result in mutational adaptations, acquisition of genetic material, or alteration of gene expression producing resistance to virtually all antibiotics currently available in clinical practice. Therefore, understanding the biochemical and genetic basis of resistance is of paramount importance to design strategies to curtail the emergence and spread of resistance and to devise innovative therapeutic approaches against multidrug-resistant organisms. In this chapter, we will describe in detail the major mechanisms of antibiotic resistance encountered in clinical practice, providing specific examples in relevant bacterial pathogens.
Figures


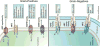

Similar articles
-
Tackling Threats and Future Problems of Multidrug-Resistant Bacteria.Curr Top Microbiol Immunol. 2016;398:3-33. doi: 10.1007/82_2016_492. Curr Top Microbiol Immunol. 2016. PMID: 27406189 Review.
-
Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance?Curr Drug Targets. 2002 Aug;3(4):345-9. doi: 10.2174/1389450023347579. Curr Drug Targets. 2002. PMID: 12102604 Review.
-
Pathogens resistant to antimicrobial agents. Epidemiology, molecular mechanisms, and clinical management.Infect Dis Clin North Am. 2000 Jun;14(2):293-319. doi: 10.1016/s0891-5520(05)70249-x. Infect Dis Clin North Am. 2000. PMID: 10829257 Review.
-
The prevention of antibiotic resistance during treatment.Infection. 1999;27 Suppl 2:S29-31. doi: 10.1007/BF02561667. Infection. 1999. PMID: 10885824 Review.
-
Antibiotic resistance in hospital pathogens--acquisition or spread?Int J Antimicrob Agents. 2001 Sep;18(3):295-8. doi: 10.1016/s0924-8579(01)00378-8. Int J Antimicrob Agents. 2001. PMID: 11673047 Review.
Cited by
-
Enabling One Health solutions through genomics.Indian J Med Res. 2021 Mar;153(3):273-279. doi: 10.4103/ijmr.IJMR_576_21. Indian J Med Res. 2021. PMID: 33906989 Free PMC article. No abstract available.
-
Terpenes as bacterial efflux pump inhibitors: A systematic review.Front Pharmacol. 2022 Oct 13;13:953982. doi: 10.3389/fphar.2022.953982. eCollection 2022. Front Pharmacol. 2022. PMID: 36313340 Free PMC article.
-
Socioeconomic factors associated with antimicrobial resistance of Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli in Chilean hospitals (2008-2017).Rev Panam Salud Publica. 2020 Sep 23;44:e30. doi: 10.26633/RPSP.2020.30. eCollection 2020. Rev Panam Salud Publica. 2020. PMID: 32973892 Free PMC article.
-
Constitutive Activation of RpoH and the Addition of L-arabinose Influence Antibiotic Sensitivity of PHL628 E. coli.Antibiotics (Basel). 2024 Feb 1;13(2):143. doi: 10.3390/antibiotics13020143. Antibiotics (Basel). 2024. PMID: 38391529 Free PMC article.
-
Expression of Anti-Lipopolysaccharide Factor Isoform 3 in Chlamydomonas reinhardtii Showing High Antimicrobial Activity.Mar Drugs. 2021 Apr 23;19(5):239. doi: 10.3390/md19050239. Mar Drugs. 2021. PMID: 33922554 Free PMC article.
References
-
- Antimicrobial resistance: global report on surveillance 2014. World Health Organization; 2014. Downloaded from http://www.who.int/drugresistance/documents/surveillancereport/en/, last accessed on March 4, 2015.
-
- Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006 Jan 15;42(Suppl 2):S82–9. - PubMed
-
- DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis. 2005 Aug 1;41(3):327–33. - PubMed
-
- Antibiotic Resistance Threats in the United States. Centers for Disease Control and Prevention. 2013 Downloaded from http://www.cdc.gov/drugresistance/threat-report-2013/index.html, last accessed on March 9, 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

