Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing
- PMID: 27189670
- PMCID: PMC4870633
- DOI: 10.1038/srep26191
Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing
Abstract
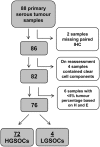
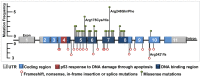
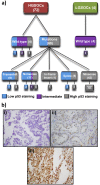
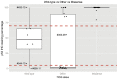
The tumour suppressor p53 is mutated in cancer, including over 96% of high-grade serous ovarian cancer (HGSOC). Mutations cause loss of wild-type p53 function due to either gain of abnormal function of mutant p53 (mutp53), or absent to low mutp53. Massively parallel sequencing (MPS) enables increased accuracy of detection of somatic variants in heterogeneous tumours. We used MPS and immunohistochemistry (IHC) to characterise HGSOCs for TP53 mutation and p53 expression. TP53 mutation was identified in 94% (68/72) of HGSOCs, 62% of which were missense. Missense mutations demonstrated high p53 by IHC, as did 35% (9/26) of non-missense mutations. Low p53 was seen by IHC in 62% of HGSOC associated with non-missense mutations. Most wild-type TP53 tumours (75%, 6/8) displayed intermediate p53 levels. The overall sensitivity of detecting a TP53 mutation based on classification as 'Low', 'Intermediate' or 'High' for p53 IHC was 99%, with a specificity of 75%. We suggest p53 IHC can be used as a surrogate marker of TP53 mutation in HGSOC; however, this will result in misclassification of a proportion of TP53 wild-type and mutant tumours. Therapeutic targeting of mutp53 will require knowledge of both TP53 mutations and mutp53 expression.
Figures



Similar articles
-
TP53 Mutation Status of Tubo-ovarian and Peritoneal High-grade Serous Carcinoma with a Wild-type p53 Immunostaining Pattern.Anticancer Res. 2017 Dec;37(12):6697-6703. doi: 10.21873/anticanres.12128. Anticancer Res. 2017. PMID: 29187446
-
Identification of Novel Somatic TP53 Mutations in Patients with High-Grade Serous Ovarian Cancer (HGSOC) Using Next-Generation Sequencing (NGS).Int J Mol Sci. 2018 May 18;19(5):1510. doi: 10.3390/ijms19051510. Int J Mol Sci. 2018. PMID: 29783665 Free PMC article.
-
Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma.J Pathol Clin Res. 2016 Jul 13;2(4):247-258. doi: 10.1002/cjp2.53. eCollection 2016 Oct. J Pathol Clin Res. 2016. PMID: 27840695 Free PMC article.
-
The Many Uses of p53 Immunohistochemistry in Gynecological Pathology: Proceedings of the ISGyP Companion Society Session at the 2020 USCAP Annual9 Meeting.Int J Gynecol Pathol. 2021 Jan;40(1):32-40. doi: 10.1097/PGP.0000000000000725. Int J Gynecol Pathol. 2021. PMID: 33290354 Review.
-
Gain-of-function mutant p53 in cancer progression and therapy.J Mol Cell Biol. 2020 Sep 1;12(9):674-687. doi: 10.1093/jmcb/mjaa040. J Mol Cell Biol. 2020. PMID: 32722796 Free PMC article. Review.
Cited by
-
Targeting lipid metabolism in the treatment of ovarian cancer.Oncotarget. 2022 May 25;13:768-783. doi: 10.18632/oncotarget.28241. eCollection 2022. Oncotarget. 2022. PMID: 35634242 Free PMC article. Review.
-
TMEFF1 overexpression and its mechanism for tumor promotion in ovarian cancer.Cancer Manag Res. 2019 Jan 17;11:839-855. doi: 10.2147/CMAR.S186080. eCollection 2019. Cancer Manag Res. 2019. PMID: 30697076 Free PMC article.
-
Discovery and development of botanical natural products and their analogues as therapeutics for ovarian cancer.Nat Prod Rep. 2023 Jul 19;40(7):1250-1270. doi: 10.1039/d2np00091a. Nat Prod Rep. 2023. PMID: 37387219 Free PMC article. Review.
-
Mutated p53 in HGSC-From a Common Mutation to a Target for Therapy.Cancers (Basel). 2021 Jul 10;13(14):3465. doi: 10.3390/cancers13143465. Cancers (Basel). 2021. PMID: 34298679 Free PMC article. Review.
-
High-grade serous carcinoma of the fallopian tube in a young woman with chromosomal 4q abnormality: A case report.World J Clin Cases. 2024 Jun 26;12(18):3539-3547. doi: 10.12998/wjcc.v12.i18.3539. World J Clin Cases. 2024. PMID: 38983400 Free PMC article.
References
-
- Soussi T. & Wiman K. G. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 12, 303–312 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

