Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies
- PMID: 27188959
- PMCID: PMC4870698
- DOI: 10.1038/srep26276
Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies
Abstract
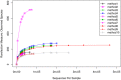
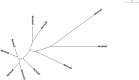
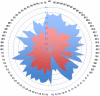
Exopolysaccharides produced by bacterial species and present in feces are extremely inhibitory to DNA restriction and can cause discrepancies in metagenomic studies. We determined the effects of different DNA extraction methods on the apparent composition of the gut microbiota using Illumina MiSeq deep sequencing technology. DNA was extracted from the stool from an obese female using 10 different methods and the choice of DNA extraction method affected the proportional abundance at the phylum level, species richness (Chao index, 227 to 2,714) and diversity (non parametric Shannon, 1.37 to 4.4). Moreover DNA was extracted from stools obtained from 83 different individuals by the fastest extraction assay and by an extraction assay that degradated exopolysaccharides. The fastest extraction method was able to detect 68% to 100% genera and 42% to 95% species whereas the glycan degradation extraction method was able to detect 56% to 93% genera and 25% to 87% species. To allow a good liberation of DNA from exopolysaccharides commonly presented in stools, we recommend the mechanical lysis of stools plus glycan degradation, used here for the first time. Caution must be taken in the interpretation of current metagenomic studies, as the efficiency of DNA extraction varies widely among stool samples.
Figures

Similar articles
-
The effect of DNA extraction methodology on gut microbiota research applications.BMC Res Notes. 2016 Jul 26;9:365. doi: 10.1186/s13104-016-2171-7. BMC Res Notes. 2016. PMID: 27456340 Free PMC article.
-
Improved yield and accuracy for DNA extraction in microbiome studies with variation in microbial biomass.Biotechniques. 2019 Jun;66(6):285-289. doi: 10.2144/btn-2019-0016. Epub 2019 May 24. Biotechniques. 2019. PMID: 31124702
-
DNA extraction for streamlined metagenomics of diverse environmental samples.Biotechniques. 2017 Jun 1;62(6):290-293. doi: 10.2144/000114559. Biotechniques. 2017. PMID: 28625159
-
Metagenomic surveys of gut microbiota.Genomics Proteomics Bioinformatics. 2015 Jun;13(3):148-58. doi: 10.1016/j.gpb.2015.02.005. Epub 2015 Jul 13. Genomics Proteomics Bioinformatics. 2015. PMID: 26184859 Free PMC article. Review.
-
The impact of intestinal microbiota on bio-medical research: definitions, techniques and physiology of a "new frontier".Acta Biomed. 2018 Dec 17;89(9-S):52-59. doi: 10.23750/abm.v89i9-S.7906. Acta Biomed. 2018. PMID: 30561396 Free PMC article. Review.
Cited by
-
Whatman Protein Saver Cards for Storage and Detection of Parasitic Enteropathogens.Am J Trop Med Hyg. 2018 Dec;99(6):1613-1618. doi: 10.4269/ajtmh.18-0538. Am J Trop Med Hyg. 2018. PMID: 30398140 Free PMC article.
-
Gut Microbiota Alteration is Characterized by a Proteobacteria and Fusobacteria Bloom in Kwashiorkor and a Bacteroidetes Paucity in Marasmus.Sci Rep. 2019 Jun 24;9(1):9084. doi: 10.1038/s41598-019-45611-3. Sci Rep. 2019. Retraction in: Sci Rep. 2023 Oct 30;13(1):18590. doi: 10.1038/s41598-023-44593-7 PMID: 31235833 Free PMC article. Retracted.
-
A Listeria monocytogenes clone in human breast milk associated with severe acute malnutrition in West Africa: A multicentric case-controlled study.PLoS Negl Trop Dis. 2021 Jun 29;15(6):e0009555. doi: 10.1371/journal.pntd.0009555. eCollection 2021 Jun. PLoS Negl Trop Dis. 2021. PMID: 34185789 Free PMC article.
-
Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals.New Microbes New Infect. 2018 Nov 2;27:14-21. doi: 10.1016/j.nmni.2018.10.009. eCollection 2019 Jan. New Microbes New Infect. 2018. PMID: 30555706 Free PMC article.
-
Metabarcoding analysis of eukaryotic microbiota in the gut of HIV-infected patients.PLoS One. 2018 Jan 31;13(1):e0191913. doi: 10.1371/journal.pone.0191913. eCollection 2018. PLoS One. 2018. PMID: 29385188 Free PMC article.
References
-
- Angelakis E., Armougom F., Million M. & Raoult D. The relationship between gut microbiota and weight gain in humans. Future. Microbiol 7, 91–109 (2012). - PubMed
-
- Angelakis E., Merhej V. & Raoult D. Related actions of probiotics and antibiotics on gut microbiota and weight modification. Lancet Infect Dis 13, 889–899 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

