Peroxisomes are platforms for cytomegalovirus' evasion from the cellular immune response
- PMID: 27181750
- PMCID: PMC4867596
- DOI: 10.1038/srep26028
Peroxisomes are platforms for cytomegalovirus' evasion from the cellular immune response
Abstract
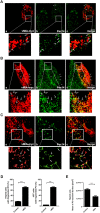
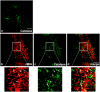
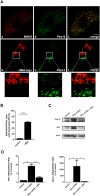
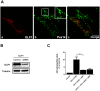
The human cytomegalovirus developed distinct evasion mechanisms from the cellular antiviral response involving vMIA, a virally-encoded protein that is not only able to prevent cellular apoptosis but also to inhibit signalling downstream from mitochondrial MAVS. vMIA has been shown to localize at mitochondria and to trigger their fragmentation, a phenomenon proven to be essential for the signalling inhibition. Here, we demonstrate that vMIA is also localized at peroxisomes, induces their fragmentation and inhibits the peroxisomal-dependent antiviral signalling pathway. Importantly, we demonstrate that peroxisomal fragmentation is not essential for vMIA to specifically inhibit signalling downstream the peroxisomal MAVS. We also show that vMIA interacts with the cytoplasmic chaperone Pex19, suggesting that the virus has developed a strategy to highjack the peroxisomal membrane proteins' transport machinery. Furthermore, we show that vMIA is able to specifically interact with the peroxisomal MAVS. Our results demonstrate that peroxisomes constitute a platform for evasion of the cellular antiviral response and that the human cytomegalovirus has developed a mechanism by which it is able to specifically evade the peroxisomal MAVS-dependent antiviral signalling.
Figures



Similar articles
-
Human Cytomegalovirus vMIA Inhibits MAVS Oligomerization at Peroxisomes in an MFF-Dependent Manner.Front Cell Dev Biol. 2022 Apr 4;10:871977. doi: 10.3389/fcell.2022.871977. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35445031 Free PMC article.
-
Hepatitis C virus NS3-4A inhibits the peroxisomal MAVS-dependent antiviral signalling response.J Cell Mol Med. 2016 Apr;20(4):750-7. doi: 10.1111/jcmm.12801. Epub 2016 Feb 10. J Cell Mol Med. 2016. PMID: 26865163 Free PMC article.
-
Herpes simplex virus 1 infection dampens the immediate early antiviral innate immunity signaling from peroxisomes by tegument protein VP16.Virol J. 2017 Feb 21;14(1):35. doi: 10.1186/s12985-017-0709-5. Virol J. 2017. PMID: 28222744 Free PMC article.
-
vMIA, a viral inhibitor of apoptosis targeting mitochondria.Biochimie. 2002 Feb-Mar;84(2-3):177-85. doi: 10.1016/s0300-9084(02)01367-6. Biochimie. 2002. PMID: 12022948 Review.
-
Superresolution imaging of viral protein trafficking.Med Microbiol Immunol. 2015 Jun;204(3):449-60. doi: 10.1007/s00430-015-0395-0. Epub 2015 Feb 28. Med Microbiol Immunol. 2015. PMID: 25724304 Free PMC article. Review.
Cited by
-
Infection-Induced Peroxisome Biogenesis Is a Metabolic Strategy for Herpesvirus Replication.Cell Host Microbe. 2018 Oct 10;24(4):526-541.e7. doi: 10.1016/j.chom.2018.09.002. Epub 2018 Sep 27. Cell Host Microbe. 2018. PMID: 30269970 Free PMC article.
-
A Cysteine Residue of Human Cytomegalovirus vMIA Protein Plays a Crucial Role in Viperin Trafficking to Control Viral Infectivity.J Virol. 2023 Jun 29;97(6):e0187422. doi: 10.1128/jvi.01874-22. Epub 2023 Jun 12. J Virol. 2023. PMID: 37306568 Free PMC article.
-
Human Cytomegalovirus vMIA Inhibits MAVS Oligomerization at Peroxisomes in an MFF-Dependent Manner.Front Cell Dev Biol. 2022 Apr 4;10:871977. doi: 10.3389/fcell.2022.871977. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35445031 Free PMC article.
-
WY14643 Increases Herpesvirus Replication and Inhibits IFNβ Production Independently of PPARα Expression.Microbiol Spectr. 2023 Jan 30;11(2):e0233722. doi: 10.1128/spectrum.02337-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36715509 Free PMC article.
-
Die Another Day: Inhibition of Cell Death Pathways by Cytomegalovirus.Viruses. 2017 Sep 2;9(9):249. doi: 10.3390/v9090249. Viruses. 2017. PMID: 28869497 Free PMC article. Review.
References
-
- Mocarski E. S., Shenk T. & Pass R. F. In Fields Virol. (Knipe D. M. & Howley P. M.) 2701–2772 (Lippincott Williams & Wilkins, 2007).
-
- Goldmacher V. S. Cell death suppression by cytomegaloviruses. Apoptosis 10, 251–65 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

