Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7
- PMID: 27171370
- PMCID: PMC4968227
- DOI: 10.1080/15548627.2016.1179401
Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7
Abstract
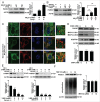
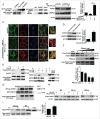
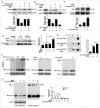
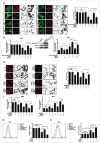
As a selective degradation system, chaperone-mediated autophagy (CMA) is essential for maintaining cellular homeostasis and survival under stress conditions. Increasing evidence points to an important role for the dysfunction of CMA in the pathogenesis of Parkinson disease (PD). However, the mechanisms by which CMA regulates neuronal survival under stress and its role in neurodegenerative diseases are not fully understood. PARK7/DJ-1 is an autosomal recessive familial PD gene. PARK7 plays a critical role in antioxidative response and its dysfunction leads to mitochondrial defects. In the current study, we showed that CMA mediated the lysosome-dependent degradation of PARK7. Importantly, CMA preferentially removed the oxidatively damaged nonfunctional PARK7 protein. Furthermore, CMA protected cells from mitochondrial toxin MPP(+)-induced changes in mitochondrial morphology and function, and increased cell viability. These protective effects were lost under PARK7-deficiency conditions. Conversely, overexpression of PARK7 significantly attenuated the mitochondrial dysfunction and cell death exacerbated by blocking CMA under oxidative stress. Thus, our findings reveal a mechanism by which CMA protects mitochondrial function by degrading nonfunctional PARK7 and maintaining its homeostasis, and dysregulation of this pathway may contribute to the neuronal stress and death in PD pathogenesis.
Keywords: CMA; DJ-1; PARK7; Parkinson disease; mitochondria; neuronal death.
Figures
Similar articles
-
Impairment of chaperone-mediated autophagy affects neuronal homeostasis through altered expression of DJ-1 and CRMP-2 proteins.Mol Cell Neurosci. 2019 Mar;95:1-12. doi: 10.1016/j.mcn.2018.12.006. Epub 2018 Dec 15. Mol Cell Neurosci. 2019. PMID: 30562574
-
Inhibitors of lysosomal function or serum starvation in control or LAMP2 deficient cells do not modify the cellular levels of Parkinson disease-associated DJ-1/PARK 7 protein.PLoS One. 2018 Jul 26;13(7):e0201152. doi: 10.1371/journal.pone.0201152. eCollection 2018. PLoS One. 2018. PMID: 30048497 Free PMC article.
-
Chaperone-mediated autophagy: roles in neuroprotection.Neurosci Bull. 2015 Aug;31(4):452-8. doi: 10.1007/s12264-015-1540-x. Epub 2015 Jul 23. Neurosci Bull. 2015. PMID: 26206599 Free PMC article. Review.
-
Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics.Hum Mol Genet. 2010 Oct 1;19(19):3734-46. doi: 10.1093/hmg/ddq288. Epub 2010 Jul 16. Hum Mol Genet. 2010. PMID: 20639397
-
Chaperone-mediated autophagy in neurodegenerative diseases: mechanisms and therapy.Mol Cell Biochem. 2023 Oct;478(10):2173-2190. doi: 10.1007/s11010-022-04640-9. Epub 2023 Jan 25. Mol Cell Biochem. 2023. PMID: 36695937 Review.
Cited by
-
Mitochondrial Degeneration and Autophagy Associated With Delayed Effects of Radiation in the Mouse Brain.Front Aging Neurosci. 2019 Dec 20;11:357. doi: 10.3389/fnagi.2019.00357. eCollection 2019. Front Aging Neurosci. 2019. PMID: 31956306 Free PMC article.
-
New Therapeutics to Modulate Mitochondrial Function in Neurodegenerative Disorders.Curr Pharm Des. 2017;23(5):731-752. doi: 10.2174/1381612822666161230144517. Curr Pharm Des. 2017. PMID: 28034353 Free PMC article. Review.
-
PARK7 deficiency inhibits fatty acid β-oxidation via PTEN to delay liver regeneration after hepatectomy.Clin Transl Med. 2022 Sep;12(9):e1061. doi: 10.1002/ctm2.1061. Clin Transl Med. 2022. PMID: 36149763 Free PMC article.
-
Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection.Front Genet. 2021 Mar 16;12:599261. doi: 10.3389/fgene.2021.599261. eCollection 2021. Front Genet. 2021. PMID: 33796130 Free PMC article.
-
The Antioxidative Role of Chaperone-Mediated Autophagy as a Downstream Regulator of Oxidative Stress in Human Diseases.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221114178. doi: 10.1177/15330338221114178. Technol Cancer Res Treat. 2022. PMID: 36131551 Free PMC article. Review.
References
-
- Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain 1991; 114:2283-301; PMID:1933245; http://dx.doi.org/10.1093/brain/114.5.2283 - DOI - PubMed
-
- Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nat Clin Pract Neurol 2008; 4:600-9; PMID:18978800; http://dx.doi.org/10.1038/ncpneuro0924 - DOI - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science 2008; 319:916-9; PMID:18276881; http://dx.doi.org/10.1126/science.1141448 - DOI - PubMed
-
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J-i, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al.. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; PMID:16625205; http://dx.doi.org/10.1038/nature04723 - DOI - PubMed
-
- He L-q, Lu J-h, Yue Z-y. Autophagy in ageing and ageing-associated diseases. Acta Pharmacol Sin 2013; 34:605-11; PMID:23416930; http://dx.doi.org/10.1038/aps.2012.188 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
