Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways
- PMID: 27164932
- PMCID: PMC4947123
- DOI: 10.1007/s00401-016-1575-8
Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways
Abstract
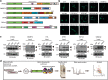
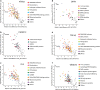
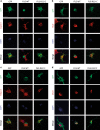
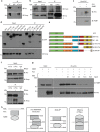
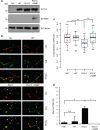
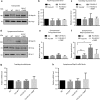
Amyotrophic lateral sclerosis (ALS) is a devastating neurological disease with no effective treatment available. An increasing number of genetic causes of ALS are being identified, but how these genetic defects lead to motor neuron degeneration and to which extent they affect common cellular pathways remains incompletely understood. To address these questions, we performed an interactomic analysis to identify binding partners of wild-type (WT) and ALS-associated mutant versions of ATXN2, C9orf72, FUS, OPTN, TDP-43 and UBQLN2 in neuronal cells. This analysis identified several known but also many novel binding partners of these proteins. Interactomes of WT and mutant ALS proteins were very similar except for OPTN and UBQLN2, in which mutations caused loss or gain of protein interactions. Several of the identified interactomes showed a high degree of overlap: shared binding partners of ATXN2, FUS and TDP-43 had roles in RNA metabolism; OPTN- and UBQLN2-interacting proteins were related to protein degradation and protein transport, and C9orf72 interactors function in mitochondria. To confirm that this overlap is important for ALS pathogenesis, we studied fragile X mental retardation protein (FMRP), one of the common interactors of ATXN2, FUS and TDP-43, in more detail in in vitro and in vivo model systems for FUS ALS. FMRP localized to mutant FUS-containing aggregates in spinal motor neurons and bound endogenous FUS in a direct and RNA-sensitive manner. Furthermore, defects in synaptic FMRP mRNA target expression, neuromuscular junction integrity, and motor behavior caused by mutant FUS in zebrafish embryos, could be rescued by exogenous FMRP expression. Together, these results show that interactomics analysis can provide crucial insight into ALS disease mechanisms and they link FMRP to motor neuron dysfunction caused by FUS mutations.
Keywords: Amyotrophic lateral sclerosis; C9orf72; FMRP; FUS; Motor neuron; TDP-43.
Figures


Similar articles
-
Human TDP-43 and FUS selectively affect motor neuron maturation and survival in a murine cell model of ALS by non-cell-autonomous mechanisms.Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(7-8):431-41. doi: 10.3109/21678421.2015.1055275. Epub 2015 Sep 7. Amyotroph Lateral Scler Frontotemporal Degener. 2015. PMID: 26174443
-
Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity.Acta Neuropathol. 2016 Apr;131(4):605-20. doi: 10.1007/s00401-015-1530-0. Epub 2016 Jan 4. Acta Neuropathol. 2016. PMID: 26728149 Free PMC article.
-
Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism.Acta Neuropathol. 2012 Nov;124(5):733-47. doi: 10.1007/s00401-012-1035-z. Epub 2012 Sep 1. Acta Neuropathol. 2012. PMID: 22941224
-
How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other?Curr Opin Neurol. 2012 Dec;25(6):701-7. doi: 10.1097/WCO.0b013e32835a269b. Curr Opin Neurol. 2012. PMID: 23041957 Review.
-
Sporadic and hereditary amyotrophic lateral sclerosis (ALS).Biochim Biophys Acta. 2015 Apr;1852(4):679-84. doi: 10.1016/j.bbadis.2014.08.010. Epub 2014 Sep 1. Biochim Biophys Acta. 2015. PMID: 25193032 Review.
Cited by
-
Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS.Biochim Biophys Acta. 2016 Oct;1862(10):2004-14. doi: 10.1016/j.bbadis.2016.07.015. Epub 2016 Jul 25. Biochim Biophys Acta. 2016. PMID: 27460707 Free PMC article.
-
Binding patterns of RNA-binding proteins to repeat-derived RNA sequences reveal putative functional RNA elements.NAR Genom Bioinform. 2021 Jul 2;3(3):lqab055. doi: 10.1093/nargab/lqab055. eCollection 2021 Sep. NAR Genom Bioinform. 2021. PMID: 34235430 Free PMC article.
-
The Fragile X Protein Family in Amyotrophic Lateral Sclerosis.Mol Neurobiol. 2023 Jul;60(7):3898-3910. doi: 10.1007/s12035-023-03330-x. Epub 2023 Mar 29. Mol Neurobiol. 2023. PMID: 36991279 Free PMC article. Review.
-
Characterization of genetic loss-of-function of Fus in zebrafish.RNA Biol. 2017 Jan 2;14(1):29-35. doi: 10.1080/15476286.2016.1256532. Epub 2016 Nov 29. RNA Biol. 2017. PMID: 27898262 Free PMC article.
-
A serum microRNA sequence reveals fragile X protein pathology in amyotrophic lateral sclerosis.Brain. 2021 May 7;144(4):1214-1229. doi: 10.1093/brain/awab018. Brain. 2021. PMID: 33871026 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

