The topography of mutational processes in breast cancer genomes
- PMID: 27136393
- PMCID: PMC5001788
- DOI: 10.1038/ncomms11383
The topography of mutational processes in breast cancer genomes
Abstract
Somatic mutations in human cancers show unevenness in genomic distribution that correlate with aspects of genome structure and function. These mutations are, however, generated by multiple mutational processes operating through the cellular lineage between the fertilized egg and the cancer cell, each composed of specific DNA damage and repair components and leaving its own characteristic mutational signature on the genome. Using somatic mutation catalogues from 560 breast cancer whole-genome sequences, here we show that each of 12 base substitution, 2 insertion/deletion (indel) and 6 rearrangement mutational signatures present in breast tissue, exhibit distinct relationships with genomic features relating to transcription, DNA replication and chromatin organization. This signature-based approach permits visualization of the genomic distribution of mutational processes associated with APOBEC enzymes, mismatch repair deficiency and homologous recombinational repair deficiency, as well as mutational processes of unknown aetiology. Furthermore, it highlights mechanistic insights including a putative replication-dependent mechanism of APOBEC-related mutagenesis.
Figures
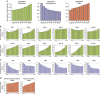

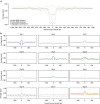

Comment in
-
Cancer genomics: A catalogue of somatic mutations.Nat Rev Genet. 2016 Jul;17(7):378. doi: 10.1038/nrg.2016.65. Epub 2016 May 9. Nat Rev Genet. 2016. PMID: 27156977 No abstract available.
Similar articles
-
Landscape of somatic mutations in 560 breast cancer whole-genome sequences.Nature. 2016 Jun 2;534(7605):47-54. doi: 10.1038/nature17676. Epub 2016 May 2. Nature. 2016. PMID: 27135926 Free PMC article.
-
Mutational signatures reveal ternary relationships between homologous recombination repair, APOBEC, and mismatch repair in gynecological cancers.J Transl Med. 2022 Feb 2;20(1):65. doi: 10.1186/s12967-022-03259-0. J Transl Med. 2022. PMID: 35109853 Free PMC article.
-
Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair.DNA Repair (Amst). 2018 Nov;71:6-11. doi: 10.1016/j.dnarep.2018.08.003. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30236628 Free PMC article. Review.
-
The repertoire of mutational signatures in human cancer.Nature. 2020 Feb;578(7793):94-101. doi: 10.1038/s41586-020-1943-3. Epub 2020 Feb 5. Nature. 2020. PMID: 32025018 Free PMC article.
-
Mechanisms underlying mutational signatures in human cancers.Nat Rev Genet. 2014 Sep;15(9):585-98. doi: 10.1038/nrg3729. Epub 2014 Jul 1. Nat Rev Genet. 2014. PMID: 24981601 Free PMC article. Review.
Cited by
-
The genetic landscape of metaplastic breast cancers and uterine carcinosarcomas.Mol Oncol. 2021 Apr;15(4):1024-1039. doi: 10.1002/1878-0261.12813. Epub 2021 Feb 19. Mol Oncol. 2021. PMID: 33021035 Free PMC article.
-
Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors.Mol Oncol. 2018 Nov;12(11):1838-1855. doi: 10.1002/1878-0261.12375. Epub 2018 Sep 21. Mol Oncol. 2018. PMID: 30133130 Free PMC article.
-
Elevated Mutational Age in Blood of Children Treated for Cancer Contributes to Therapy-Related Myeloid Neoplasms.Cancer Discov. 2022 Aug 5;12(8):1860-1872. doi: 10.1158/2159-8290.CD-22-0120. Cancer Discov. 2022. PMID: 35678530 Free PMC article.
-
The Rad51 paralogs facilitate a novel DNA strand specific damage tolerance pathway.Nat Commun. 2019 Aug 5;10(1):3515. doi: 10.1038/s41467-019-11374-8. Nat Commun. 2019. PMID: 31383866 Free PMC article.
-
Single-stranded DNA binding proteins influence APOBEC3A substrate preference.Sci Rep. 2021 Oct 25;11(1):21008. doi: 10.1038/s41598-021-00435-y. Sci Rep. 2021. PMID: 34697369 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

