Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity
- PMID: 27133168
- PMCID: PMC4889222
- DOI: 10.1016/j.cell.2016.04.012
Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity
Abstract
Across eukaryotic species, mild mitochondrial stress can have beneficial effects on the lifespan of organisms. Mitochondrial dysfunction activates an unfolded protein response (UPR(mt)), a stress signaling mechanism designed to ensure mitochondrial homeostasis. Perturbation of mitochondria during larval development in C. elegans not only delays aging but also maintains UPR(mt) signaling, suggesting an epigenetic mechanism that modulates both longevity and mitochondrial proteostasis throughout life. We identify the conserved histone lysine demethylases jmjd-1.2/PHF8 and jmjd-3.1/JMJD3 as positive regulators of lifespan in response to mitochondrial dysfunction across species. Reduction of function of the demethylases potently suppresses longevity and UPR(mt) induction, while gain of function is sufficient to extend lifespan in a UPR(mt)-dependent manner. A systems genetics approach in the BXD mouse reference population further indicates conserved roles of the mammalian orthologs in longevity and UPR(mt) signaling. These findings illustrate an evolutionary conserved epigenetic mechanism that determines the rate of aging downstream of mitochondrial perturbations.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
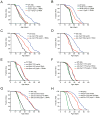
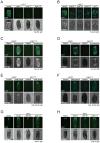
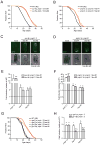
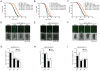
Comment in
-
Mitochondria: Masters of Epigenetics.Cell. 2016 May 19;165(5):1052-1054. doi: 10.1016/j.cell.2016.05.021. Cell. 2016. PMID: 27203109 Free PMC article.
-
Ageing: The yin and yang of mitochondrial dysfunction.Nat Rev Mol Cell Biol. 2016 May 23;17(6):331. doi: 10.1038/nrm.2016.71. Nat Rev Mol Cell Biol. 2016. PMID: 27211483 No abstract available.
Similar articles
-
The transcriptional coactivator CBP/p300 is an evolutionarily conserved node that promotes longevity in response to mitochondrial stress.Nat Aging. 2021 Feb;1(2):165-178. doi: 10.1038/s43587-020-00025-z. Epub 2021 Feb 8. Nat Aging. 2021. PMID: 33718883 Free PMC article.
-
H3K27 modifiers regulate lifespan in C. elegans in a context-dependent manner.BMC Biol. 2021 Mar 25;19(1):59. doi: 10.1186/s12915-021-00984-8. BMC Biol. 2021. PMID: 33766022 Free PMC article.
-
Physiology: Stressed-out chromatin promotes longevity.Nature. 2016 Jun 30;534(7609):625-6. doi: 10.1038/534625a. Nature. 2016. PMID: 27357789 Free PMC article.
-
Mitochondrial stress: balancing friend and foe.Exp Gerontol. 2014 Aug;56:194-201. doi: 10.1016/j.exger.2014.02.013. Epub 2014 Mar 3. Exp Gerontol. 2014. PMID: 24603155 Review.
-
Sirtuins and the Estrogen Receptor as Regulators of the Mammalian Mitochondrial UPR in Cancer and Aging.Adv Cancer Res. 2016;130:211-56. doi: 10.1016/bs.acr.2016.01.004. Epub 2016 Mar 2. Adv Cancer Res. 2016. PMID: 27037754 Review.
Cited by
-
Understanding the Role of the SMN Complex Component GEMIN5 and Its Functional Relationship with Demethylase KDM6B in the Flunarizine-Mediated Neuroprotection of Motor Neuron Disease Spinal Muscular Atrophy.Int J Mol Sci. 2024 Sep 18;25(18):10039. doi: 10.3390/ijms251810039. Int J Mol Sci. 2024. PMID: 39337533 Free PMC article.
-
Genetic and Epigenetic Interactions Involved in Senescence of Stem Cells.Int J Mol Sci. 2024 Sep 7;25(17):9708. doi: 10.3390/ijms25179708. Int J Mol Sci. 2024. PMID: 39273655 Free PMC article. Review.
-
A bird's eye view of mitochondrial unfolded protein response in cancer: mechanisms, progression and further applications.Cell Death Dis. 2024 Sep 11;15(9):667. doi: 10.1038/s41419-024-07049-y. Cell Death Dis. 2024. PMID: 39261452 Free PMC article. Review.
-
Mitochondria: fundamental characteristics, challenges, and impact on aging.Biogerontology. 2024 Nov;25(6):923-941. doi: 10.1007/s10522-024-10132-8. Epub 2024 Aug 28. Biogerontology. 2024. PMID: 39196438 Review.
-
Versatile JMJD proteins: juggling histones and much more.Trends Biochem Sci. 2024 Sep;49(9):804-818. doi: 10.1016/j.tibs.2024.06.009. Epub 2024 Jun 26. Trends Biochem Sci. 2024. PMID: 38926050 Review.
References
-
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
-
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol CB. 2004;14:1639–1643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

