Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells
- PMID: 27129276
- PMCID: PMC4933233
- DOI: 10.1074/jbc.M115.700534
Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells
Abstract
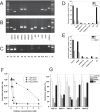
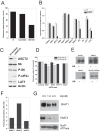
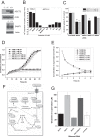
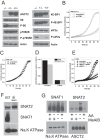
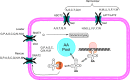
Many cancer cells depend on glutamine as they use the glutaminolysis pathway to generate building blocks and energy for anabolic purposes. As a result, glutamine transporters are essential for cancer growth and are potential targets for cancer chemotherapy with ASCT2 (SLC1A5) being investigated most intensively. Here we show that HeLa epithelial cervical cancer cells and 143B osteosarcoma cells express a set of glutamine transporters including SNAT1 (SLC38A1), SNAT2 (SLC38A2), SNAT4 (SLC38A4), LAT1 (SLC7A5), and ASCT2 (SLC1A5). Net glutamine uptake did not depend on ASCT2 but required expression of SNAT1 and SNAT2. Deletion of ASCT2 did not reduce cell growth but caused an amino acid starvation response and up-regulation of SNAT1 to replace ASCT2 functionally. Silencing of GCN2 in the ASCT2(-/-) background reduced cell growth, showing that a combined targeted approach would inhibit growth of glutamine-dependent cancer cells.
Keywords: amino acid transport; cell growth; glutamine; mammalian target of rapamycin (mTOR); membrane transport.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells.J Biol Chem. 2019 Mar 15;294(11):4012-4026. doi: 10.1074/jbc.RA118.006378. Epub 2019 Jan 11. J Biol Chem. 2019. PMID: 30635397 Free PMC article.
-
The glutamine transporter ASCT2 (SLC1A5) promotes tumor growth independently of the amino acid transporter LAT1 (SLC7A5).J Biol Chem. 2018 Feb 23;293(8):2877-2887. doi: 10.1074/jbc.RA117.001342. Epub 2018 Jan 11. J Biol Chem. 2018. PMID: 29326164 Free PMC article.
-
Disruption of Amino Acid Homeostasis by Novel ASCT2 Inhibitors Involves Multiple Targets.Front Pharmacol. 2018 Jul 19;9:785. doi: 10.3389/fphar.2018.00785. eCollection 2018. Front Pharmacol. 2018. PMID: 30072900 Free PMC article.
-
The role of the glutamine transporter ASCT2 in antineoplastic therapy.Cancer Chemother Pharmacol. 2021 Apr;87(4):447-464. doi: 10.1007/s00280-020-04218-6. Epub 2021 Jan 19. Cancer Chemother Pharmacol. 2021. PMID: 33464409 Review.
-
The transport of glutamine into mammalian cells.Front Biosci. 2007 Jan 1;12:874-82. doi: 10.2741/2109. Front Biosci. 2007. PMID: 17127344 Review.
Cited by
-
Transporters at the Interface between Cytosolic and Mitochondrial Amino Acid Metabolism.Metabolites. 2021 Feb 16;11(2):112. doi: 10.3390/metabo11020112. Metabolites. 2021. PMID: 33669382 Free PMC article. Review.
-
The Interaction between Collagen 1 and High Mannose Type CD133 Up-Regulates Glutamine Transporter SLC1A5 to Promote the Tumorigenesis of Glioblastoma Stem Cells.Adv Sci (Weinh). 2024 Jan;11(3):e2306715. doi: 10.1002/advs.202306715. Epub 2023 Nov 23. Adv Sci (Weinh). 2024. PMID: 37997289 Free PMC article.
-
Retromer and TBC1D5 maintain late endosomal RAB7 domains to enable amino acid-induced mTORC1 signaling.J Cell Biol. 2019 Sep 2;218(9):3019-3038. doi: 10.1083/jcb.201812110. Epub 2019 Aug 20. J Cell Biol. 2019. PMID: 31431476 Free PMC article.
-
Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells.J Biol Chem. 2019 Mar 15;294(11):4012-4026. doi: 10.1074/jbc.RA118.006378. Epub 2019 Jan 11. J Biol Chem. 2019. PMID: 30635397 Free PMC article.
-
Tumor necrosis factor induces pathogenic mitochondrial ROS in tuberculosis through reverse electron transport.Science. 2022 Jun 24;376(6600):eabh2841. doi: 10.1126/science.abh2841. Epub 2022 Jun 24. Science. 2022. PMID: 35737799 Free PMC article.
References
-
- Chandel N. S. (2015) Navigating Metabolism, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Newsholme E. A., Crabtree B., and Ardawi M. S. (1985) Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q. J. Exp. Physiol. 70, 473–489 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

