Zinc Finger Protein 618 Regulates the Function of UHRF2 (Ubiquitin-like with PHD and Ring Finger Domains 2) as a Specific 5-Hydroxymethylcytosine Reader
- PMID: 27129234
- PMCID: PMC4919451
- DOI: 10.1074/jbc.M116.717314
Zinc Finger Protein 618 Regulates the Function of UHRF2 (Ubiquitin-like with PHD and Ring Finger Domains 2) as a Specific 5-Hydroxymethylcytosine Reader
Abstract
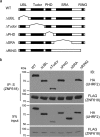
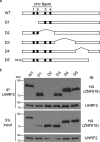
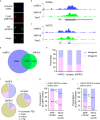
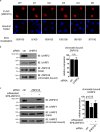
5-Hydroxymethylcytosine (5hmC) is an epigenetic modification that is generated by ten-eleven translocation (TET) protein-mediated oxidation of 5-methylcytosine (5mC). 5hmC is associated with transcription regulation and is decreased in many cancers including melanoma. Accumulating evidence has suggested that 5hmC is functionally distinct from 5mC. Ubiquitin-like with PHD and ring finger domains 2 (UHRF2) is the first known specific 5hmC reader that has higher affinity to 5hmC than 5mC, suggesting that UHRF2 might mediate 5hmC's function. Structural analysis has revealed the molecular mechanism of UHRF2-5hmC binding in vitro, but it is not clear how UHRF2 recognizes 5hmC in vivo In this study, we have identified zinc figure protein 618 (ZNF618) as a novel binding partner of UHRF2. ZNF618 specifically interacts with UHRF2 but not its paralog UHRF1. Importantly, ZNF618 co-localizes with UHRF2 at genomic loci that are enriched for 5hmC. The ZNF618 chromatin localization is independent of its interaction with UHRF2 and is through its first two zinc fingers. Instead, ZNF618 regulates UHRF2 chromatin localization. Collectively, our study suggests that ZNF618 is a key protein that regulates UHRF2 function as a specific 5hmC reader in vivo.
Keywords: 5-hydroxymethylcytosine (5-hmC); DNA demethylation; chromatin; molecular biology; molecular cell biology.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Loss of UHRF2 expression is associated with human neoplasia, promoter hypermethylation, decreased 5-hydroxymethylcytosine, and high proliferative activity.Oncotarget. 2016 Nov 15;7(46):76047-76061. doi: 10.18632/oncotarget.12583. Oncotarget. 2016. PMID: 27738314 Free PMC article.
-
Demethylation Status of Somatic DNA Extracted From Pituitary Neuroendocrine Tumors Indicates Proliferative Behavior.J Clin Endocrinol Metab. 2020 Jun 1;105(6):dgaa156. doi: 10.1210/clinem/dgaa156. J Clin Endocrinol Metab. 2020. PMID: 32232382
-
UHRF2 commissions the completion of DNA demethylation through allosteric activation by 5hmC and K33-linked ubiquitination of XRCC1.Mol Cell. 2021 Jul 15;81(14):2960-2974.e7. doi: 10.1016/j.molcel.2021.05.022. Epub 2021 Jun 9. Mol Cell. 2021. PMID: 34111398
-
5-Hydroxymethylcytosine: a stable or transient DNA modification?Genomics. 2014 Nov;104(5):314-23. doi: 10.1016/j.ygeno.2014.08.015. Epub 2014 Aug 30. Genomics. 2014. PMID: 25181633 Free PMC article. Review.
-
Genomic distribution and possible functions of DNA hydroxymethylation in the brain.Genomics. 2014 Nov;104(5):341-6. doi: 10.1016/j.ygeno.2014.08.020. Epub 2014 Sep 7. Genomics. 2014. PMID: 25205307 Review.
Cited by
-
DNA Methylation and Hydroxymethylation in Primary Colon Cancer and Synchronous Hepatic Metastasis.Front Genet. 2018 Jan 9;8:229. doi: 10.3389/fgene.2017.00229. eCollection 2017. Front Genet. 2018. PMID: 29375619 Free PMC article.
-
Overexpression of UHRF2 in intrahepatic cholangiocarcinoma and its clinical significance.Onco Targets Ther. 2017 Dec 11;10:5863-5872. doi: 10.2147/OTT.S149361. eCollection 2017. Onco Targets Ther. 2017. PMID: 29270024 Free PMC article.
-
Loss of UHRF2 expression is associated with human neoplasia, promoter hypermethylation, decreased 5-hydroxymethylcytosine, and high proliferative activity.Oncotarget. 2016 Nov 15;7(46):76047-76061. doi: 10.18632/oncotarget.12583. Oncotarget. 2016. PMID: 27738314 Free PMC article.
-
DiPRO1 distinctly reprograms muscle and mesenchymal cancer cells.EMBO Mol Med. 2024 Aug;16(8):1840-1885. doi: 10.1038/s44321-024-00097-z. Epub 2024 Jul 15. EMBO Mol Med. 2024. PMID: 39009887 Free PMC article.
-
Single cell RNA-seq reveals genes vital to in vitro fertilized embryos and parthenotes in pigs.Sci Rep. 2021 Jul 13;11(1):14393. doi: 10.1038/s41598-021-93904-3. Sci Rep. 2021. PMID: 34257377 Free PMC article.
References
-
- Li E., Bestor T. H., and Jaenisch R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

