Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation
- PMID: 27127234
- PMCID: PMC5193222
- DOI: 10.1126/science.aad7993
Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation
Abstract
The host innate immune response is the first line of defense against pathogens and is orchestrated by the concerted expression of genes induced by microbial stimuli. Deregulated expression of these genes is linked to the initiation and progression of diseases associated with exacerbated inflammation. We identified topoisomerase 1 (Top1) as a positive regulator of RNA polymerase II transcriptional activity at pathogen-induced genes. Depletion or chemical inhibition of Top1 suppresses the host response against influenza and Ebola viruses as well as bacterial products. Therapeutic pharmacological inhibition of Top1 protected mice from death in experimental models of lethal inflammation. Our results indicate that Top1 inhibition could be used as therapy against life-threatening infections characterized by an acutely exacerbated immune response.
Copyright © 2016, American Association for the Advancement of Science.
Figures



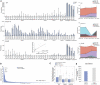


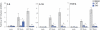
Comment in
-
GENE EXPRESSION. Unwinding inducible gene expression.Science. 2016 May 27;352(6289):1058-9. doi: 10.1126/science.aag0365. Science. 2016. PMID: 27230367 No abstract available.
Similar articles
-
GENE EXPRESSION. Unwinding inducible gene expression.Science. 2016 May 27;352(6289):1058-9. doi: 10.1126/science.aag0365. Science. 2016. PMID: 27230367 No abstract available.
-
TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation.Cell. 2021 May 13;184(10):2618-2632.e17. doi: 10.1016/j.cell.2021.03.051. Epub 2021 Mar 30. Cell. 2021. PMID: 33836156 Free PMC article.
-
Topoisomerase 1 Inhibition Promotes Cyclic GMP-AMP Synthase-Dependent Antiviral Responses.mBio. 2017 Oct 3;8(5):e01611-17. doi: 10.1128/mBio.01611-17. mBio. 2017. PMID: 28974621 Free PMC article.
-
Dissecting the transcriptional functions of human DNA topoisomerase I by selective inhibitors: implications for physiological and therapeutic modulation of enzyme activity.Biochim Biophys Acta. 2010 Dec;1806(2):240-50. doi: 10.1016/j.bbcan.2010.06.003. Epub 2010 Jun 27. Biochim Biophys Acta. 2010. PMID: 20600630 Review.
-
[Molecular determinants of response to topoisomerase I inhibitors].Bull Cancer. 2011 Nov;98(11):1287-98. doi: 10.1684/bdc.2011.1474. Bull Cancer. 2011. PMID: 22049384 Review. French.
Cited by
-
Development of Novel 1,3-Disubstituted-2-Thiohydantoin Analogues with Potent Anti-Inflammatory Activity; In Vitro and In Silico Assessments.Molecules. 2022 Sep 23;27(19):6271. doi: 10.3390/molecules27196271. Molecules. 2022. PMID: 36234810 Free PMC article.
-
The Natural Compound Neobractatin Induces Cell Cycle Arrest by Regulating E2F1 and Gadd45α.Front Oncol. 2019 Jul 17;9:654. doi: 10.3389/fonc.2019.00654. eCollection 2019. Front Oncol. 2019. PMID: 31380287 Free PMC article.
-
Myeloid cell-specific topoisomerase 1 inhibition using DNA origami mitigates neuroinflammation.EMBO Rep. 2022 Jul 5;23(7):e54499. doi: 10.15252/embr.202154499. Epub 2022 May 20. EMBO Rep. 2022. PMID: 35593064 Free PMC article.
-
MicroRNA-155 attenuates late sepsis-induced cardiac dysfunction through JNK and β-arrestin 2.Oncotarget. 2017 Jul 18;8(29):47317-47329. doi: 10.18632/oncotarget.17636. Oncotarget. 2017. PMID: 28525390 Free PMC article.
-
An in silico analysis identifies drugs potentially modulating the cytokine storm triggered by SARS-CoV-2 infection.Sci Rep. 2022 Jan 31;12(1):1626. doi: 10.1038/s41598-022-05597-x. Sci Rep. 2022. PMID: 35102208 Free PMC article.
References
-
- Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359; pmid: 11861602. - PubMed
-
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004; pmid: 19538928. - PubMed
-
- Beutler B, et al. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 2007;7:753–766. doi: 10.1038/nri2174; pmid: 17893693. - PubMed
-
- Crow YJ. Type I interferonopathies: Mendelian type I interferon up-regulation. Curr. Opin. Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005; pmid: 25463593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007647/AI/NIAID NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- R01 AI113186/AI/NIAID NIH HHS/United States
- R01 CA154947/CA/NCI NIH HHS/United States
- U19AI109664/AI/NIAID NIH HHS/United States
- 1R56AI114770-01A1/AI/NIAID NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- HHSN272201400008C/PHS HHS/United States
- R56 AI114770/AI/NIAID NIH HHS/United States
- 1R01AN3663134/PHS HHS/United States
- U19AI106754/AI/NIAID NIH HHS/United States
- AI07647/AI/NIAID NIH HHS/United States
- U19 AI109945/AI/NIAID NIH HHS/United States
- U19 AI106754/AI/NIAID NIH HHS/United States
- U19AI109945/AI/NIAID NIH HHS/United States
- U19 AI109664/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

