Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer's Patch T Follicular Helper Cells
- PMID: 27096318
- PMCID: PMC5296410
- DOI: 10.1016/j.immuni.2016.03.013
Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer's Patch T Follicular Helper Cells
Abstract
Gut microbiota profoundly affect gut and systemic diseases, but the mechanism whereby microbiota affect systemic diseases is unclear. It is not known whether specific microbiota regulate T follicular helper (Tfh) cells, whose excessive responses can inflict antibody-mediated autoimmunity. Using the K/BxN autoimmune arthritis model, we demonstrated that Peyer's patch (PP) Tfh cells were essential for gut commensal segmented filamentous bacteria (SFB)-induced systemic arthritis despite the production of auto-antibodies predominantly occurring in systemic lymphoid tissues, not PPs. We determined that SFB, by driving differentiation and egress of PP Tfh cells into systemic sites, boosted systemic Tfh cell and auto-antibody responses that exacerbated arthritis. SFB induced PP Tfh cell differentiation by limiting the access of interleukin 2 to CD4(+) T cells, thereby enhancing Tfh cell master regulator Bcl-6 in a dendritic cell-dependent manner. These findings showed that gut microbiota remotely regulated a systemic disease by driving the induction and egress of gut Tfh cells.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
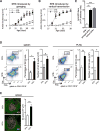

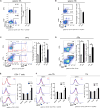
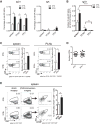
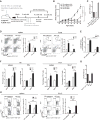

Similar articles
-
P2RX7 Deletion in T Cells Promotes Autoimmune Arthritis by Unleashing the Tfh Cell Response.Front Immunol. 2019 Mar 19;10:411. doi: 10.3389/fimmu.2019.00411. eCollection 2019. Front Immunol. 2019. PMID: 30949163 Free PMC article.
-
The impact of age and gut microbiota on Th17 and Tfh cells in K/BxN autoimmune arthritis.Arthritis Res Ther. 2017 Aug 15;19(1):188. doi: 10.1186/s13075-017-1398-6. Arthritis Res Ther. 2017. PMID: 28810929 Free PMC article.
-
Gut Commensal Segmented Filamentous Bacteria Fine-Tune T Follicular Regulatory Cells to Modify the Severity of Systemic Autoimmune Arthritis.J Immunol. 2021 Mar 1;206(5):941-952. doi: 10.4049/jimmunol.2000663. Epub 2021 Jan 18. J Immunol. 2021. PMID: 33462137 Free PMC article.
-
[T follicular helper (Tfh) cells in autoimmune diseases].Nihon Rinsho Meneki Gakkai Kaishi. 2016;39(1):1-7. doi: 10.2177/jsci.39.1. Nihon Rinsho Meneki Gakkai Kaishi. 2016. PMID: 27181228 Review. Japanese.
-
The Roles of Peyer's Patches and Microfold Cells in the Gut Immune System: Relevance to Autoimmune Diseases.Front Immunol. 2019 Oct 9;10:2345. doi: 10.3389/fimmu.2019.02345. eCollection 2019. Front Immunol. 2019. PMID: 31649668 Free PMC article. Review.
Cited by
-
Microbiota activation and regulation of adaptive immunity.Front Immunol. 2024 Oct 9;15:1429436. doi: 10.3389/fimmu.2024.1429436. eCollection 2024. Front Immunol. 2024. PMID: 39445008 Free PMC article. Review.
-
The Potential Role of Butyrate in the Pathogenesis and Treatment of Autoimmune Rheumatic Diseases.Biomedicines. 2024 Aug 5;12(8):1760. doi: 10.3390/biomedicines12081760. Biomedicines. 2024. PMID: 39200224 Free PMC article. Review.
-
The aberrant tonsillar microbiota modulates autoimmune responses in rheumatoid arthritis.JCI Insight. 2024 Aug 20;9(18):e175916. doi: 10.1172/jci.insight.175916. JCI Insight. 2024. PMID: 39163137 Free PMC article.
-
Critical role of the gut microbiota in immune responses and cancer immunotherapy.J Hematol Oncol. 2024 May 14;17(1):33. doi: 10.1186/s13045-024-01541-w. J Hematol Oncol. 2024. PMID: 38745196 Free PMC article. Review.
-
Intestinal microbiota programming of alveolar macrophages influences severity of respiratory viral infection.Cell Host Microbe. 2024 Mar 13;32(3):335-348.e8. doi: 10.1016/j.chom.2024.01.002. Epub 2024 Jan 30. Cell Host Microbe. 2024. PMID: 38295788
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

