Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity
- PMID: 27091476
- PMCID: PMC4834828
- DOI: 10.1186/s13059-016-0944-x
Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity
Abstract
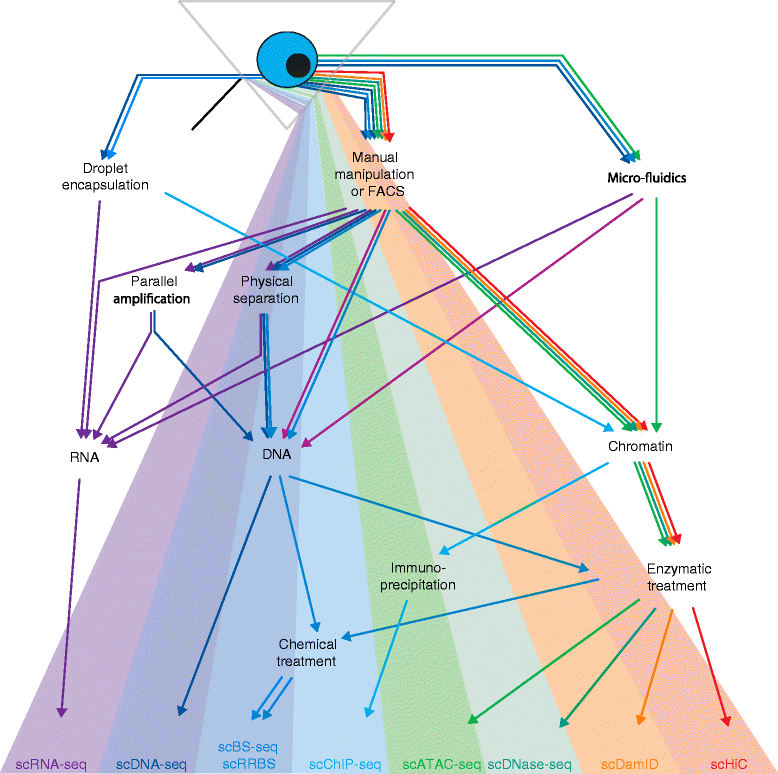
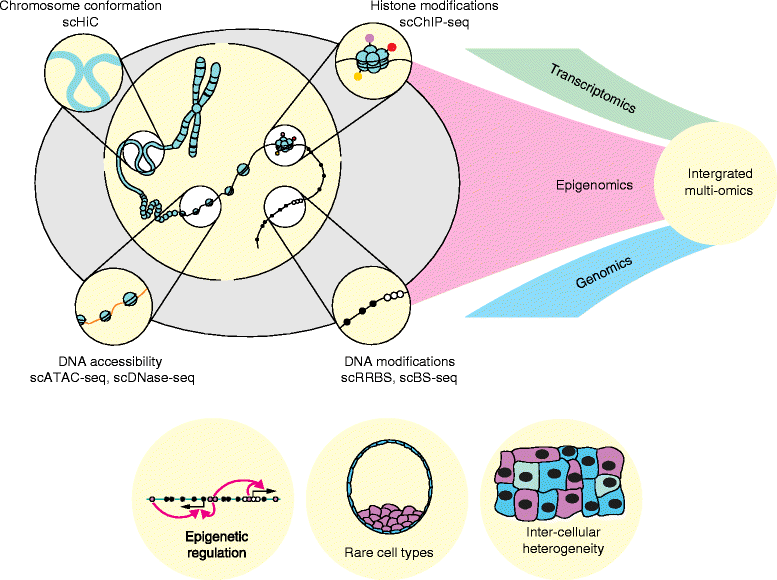
Emerging single-cell epigenomic methods are being developed with the exciting potential to transform our knowledge of gene regulation. Here we review available techniques and future possibilities, arguing that the full potential of single-cell epigenetic studies will be realized through parallel profiling of genomic, transcriptional, and epigenetic information.
Figures
Similar articles
-
Single-cell epigenomics: Recording the past and predicting the future.Science. 2017 Oct 6;358(6359):69-75. doi: 10.1126/science.aan6826. Science. 2017. PMID: 28983045 Review.
-
Single-cell epigenomics: techniques and emerging applications.Nat Rev Genet. 2015 Dec;16(12):716-26. doi: 10.1038/nrg3980. Epub 2015 Oct 13. Nat Rev Genet. 2015. PMID: 26460349 Review.
-
scMET: Bayesian modeling of DNA methylation heterogeneity at single-cell resolution.Genome Biol. 2021 Apr 20;22(1):114. doi: 10.1186/s13059-021-02329-8. Genome Biol. 2021. PMID: 33879195 Free PMC article.
-
Epigenomics in the single cell era, an important read out for genome function and cell identity.Epigenomics. 2021 Jul;13(13):981-984. doi: 10.2217/epi-2021-0153. Epub 2021 Jun 11. Epigenomics. 2021. PMID: 34114476 No abstract available.
-
Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution.Nat Genet. 2019 Jan;51(1):19-25. doi: 10.1038/s41588-018-0290-x. Epub 2018 Dec 17. Nat Genet. 2019. PMID: 30559489 Review.
Cited by
-
Opportunities and Challenges in Advancing Plant Research with Single-cell Omics.Genomics Proteomics Bioinformatics. 2024 Jul 3;22(2):qzae026. doi: 10.1093/gpbjnl/qzae026. Genomics Proteomics Bioinformatics. 2024. PMID: 38996445 Free PMC article. Review.
-
Network Approaches for Dissecting the Immune System.iScience. 2020 Aug 21;23(8):101354. doi: 10.1016/j.isci.2020.101354. Epub 2020 Jul 10. iScience. 2020. PMID: 32717640 Free PMC article. Review.
-
Joint reconstruction of cis-regulatory interaction networks across multiple tissues using single-cell chromatin accessibility data.Brief Bioinform. 2021 May 20;22(3):bbaa120. doi: 10.1093/bib/bbaa120. Brief Bioinform. 2021. PMID: 32578841 Free PMC article.
-
The Kaleidoscope of Microglial Phenotypes.Front Immunol. 2018 Jul 31;9:1753. doi: 10.3389/fimmu.2018.01753. eCollection 2018. Front Immunol. 2018. PMID: 30108586 Free PMC article. Review.
-
DNA methylation clocks tick in naked mole rats but queens age more slowly than nonbreeders.Nat Aging. 2022 Jan;2(1):46-59. doi: 10.1038/s43587-021-00152-1. Epub 2021 Dec 23. Nat Aging. 2022. PMID: 35368774 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

