HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity
- PMID: 27067600
- PMCID: PMC4858568
- DOI: 10.1016/j.molcel.2016.03.008
HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity
Abstract
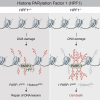
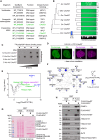
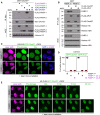
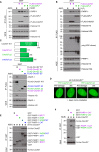
We report the identification of histone PARylation factor 1 (HPF1; also known as C4orf27) as a regulator of ADP-ribosylation signaling in the DNA damage response. HPF1/C4orf27 forms a robust protein complex with PARP-1 in cells and is recruited to DNA lesions in a PARP-1-dependent manner, but independently of PARP-1 catalytic ADP-ribosylation activity. Functionally, HPF1 promotes PARP-1-dependent in trans ADP-ribosylation of histones and limits DNA damage-induced hyper-automodification of PARP-1. Human cells lacking HPF1 exhibit sensitivity to DNA damaging agents and PARP inhibition, thereby suggesting an important role for HPF1 in genome maintenance and regulating the efficacy of PARP inhibitors. Collectively, our results demonstrate how a fundamental step in PARP-1-dependent ADP-ribosylation signaling is regulated and suggest that HPF1 functions at the crossroads of histone ADP-ribosylation and PARP-1 automodification.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Serine ADP-Ribosylation Depends on HPF1.Mol Cell. 2017 Mar 2;65(5):932-940.e6. doi: 10.1016/j.molcel.2017.01.003. Epub 2017 Feb 9. Mol Cell. 2017. PMID: 28190768 Free PMC article.
-
HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.Nature. 2020 Mar;579(7800):598-602. doi: 10.1038/s41586-020-2013-6. Epub 2020 Feb 6. Nature. 2020. PMID: 32028527 Free PMC article.
-
[Poly(ADP-Ribose) Polymerases 1 and 2: Classical Functions and Interaction with New Histone Poly(ADP-Ribosyl)ation Factor HPF1].Mol Biol (Mosk). 2023 Mar-Apr;57(2):254-268. Mol Biol (Mosk). 2023. PMID: 37000654 Russian.
-
PARP Power: A Structural Perspective on PARP1, PARP2, and PARP3 in DNA Damage Repair and Nucleosome Remodelling.Int J Mol Sci. 2021 May 12;22(10):5112. doi: 10.3390/ijms22105112. Int J Mol Sci. 2021. PMID: 34066057 Free PMC article. Review.
-
ADP-ribose contributions to genome stability and PARP enzyme trapping on sites of DNA damage; paradigm shifts for a coming-of-age modification.J Biol Chem. 2023 Dec;299(12):105397. doi: 10.1016/j.jbc.2023.105397. Epub 2023 Oct 28. J Biol Chem. 2023. PMID: 37898399 Free PMC article. Review.
Cited by
-
Role of PARP-1 in Human Cytomegalovirus Infection and Functional Partners Encoded by This Virus.Viruses. 2022 Sep 15;14(9):2049. doi: 10.3390/v14092049. Viruses. 2022. PMID: 36146855 Free PMC article.
-
Updated protein domain annotation of the PARP protein family sheds new light on biological function.Nucleic Acids Res. 2023 Aug 25;51(15):8217-8236. doi: 10.1093/nar/gkad514. Nucleic Acids Res. 2023. PMID: 37326024 Free PMC article.
-
PARP1 and HPF1 team up to flag down DNA-repair machinery.Nat Struct Mol Biol. 2023 May;30(5):568-569. doi: 10.1038/s41594-023-00987-9. Nat Struct Mol Biol. 2023. PMID: 37161004 No abstract available.
-
HPF1-dependent histone ADP-ribosylation triggers chromatin relaxation to promote the recruitment of repair factors at sites of DNA damage.Nat Struct Mol Biol. 2023 May;30(5):678-691. doi: 10.1038/s41594-023-00977-x. Epub 2023 Apr 27. Nat Struct Mol Biol. 2023. PMID: 37106138
-
The function and regulation of ADP-ribosylation in the DNA damage response.Biochem Soc Trans. 2023 Jun 28;51(3):995-1008. doi: 10.1042/BST20220749. Biochem Soc Trans. 2023. PMID: 37171085 Free PMC article.
References
-
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. - PubMed
-
- Barkauskaite E., Jankevicius G., Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell. 2015;58:935–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

