CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice
- PMID: 27066908
- PMCID: PMC4828655
- DOI: 10.1038/srep24250
CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice
Abstract
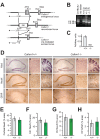
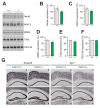
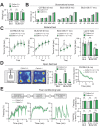
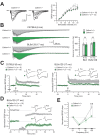
CALHM1 is a cell surface calcium channel expressed in cerebral neurons. CALHM1 function in the brain remains unknown, but recent results showed that neuronal CALHM1 controls intracellular calcium signaling and cell excitability, two mechanisms required for synaptic function. Here, we describe the generation of Calhm1 knockout (Calhm1(-/-)) mice and investigate CALHM1 role in neuronal and cognitive functions. Structural analysis revealed that Calhm1(-/-) brains had normal regional and cellular architecture, and showed no evidence of neuronal or synaptic loss, indicating that CALHM1 deficiency does not affect brain development or brain integrity in adulthood. However, Calhm1(-/-) mice showed a severe impairment in memory flexibility, assessed in the Morris water maze, and a significant disruption of long-term potentiation without alteration of long-term depression, measured in ex vivo hippocampal slices. Importantly, in primary neurons and hippocampal slices, CALHM1 activation facilitated the phosphorylation of NMDA and AMPA receptors by protein kinase A. Furthermore, neuronal CALHM1 activation potentiated the effect of glutamate on the expression of c-Fos and C/EBPβ, two immediate-early gene markers of neuronal activity. Thus, CALHM1 controls synaptic activity in cerebral neurons and is required for the flexible processing of memory in mice. These results shed light on CALHM1 physiology in the mammalian brain.
Figures

Similar articles
-
CALHM1 controls the Ca²⁺-dependent MEK, ERK, RSK and MSK signaling cascade in neurons.J Cell Sci. 2013 Mar 1;126(Pt 5):1199-206. doi: 10.1242/jcs.117135. Epub 2013 Jan 23. J Cell Sci. 2013. PMID: 23345406 Free PMC article.
-
CALHM1 ion channel elicits amyloid-β clearance by insulin-degrading enzyme in cell lines and in vivo in the mouse brain.J Cell Sci. 2015 Jul 1;128(13):2330-8. doi: 10.1242/jcs.167270. Epub 2015 May 21. J Cell Sci. 2015. PMID: 25999473 Free PMC article.
-
Post-translational palmitoylation controls the voltage gating and lipid raft association of the CALHM1 channel.J Physiol. 2017 Sep 15;595(18):6121-6145. doi: 10.1113/JP274164. Epub 2017 Aug 14. J Physiol. 2017. PMID: 28734079 Free PMC article.
-
Auxiliary subunits assist AMPA-type glutamate receptors.Science. 2006 Mar 3;311(5765):1253-6. doi: 10.1126/science.1123339. Science. 2006. PMID: 16513974 Review.
-
[Molecular mechanisms for memory formation].Brain Nerve. 2008 Jul;60(7):707-15. Brain Nerve. 2008. PMID: 18646610 Review. Japanese.
Cited by
-
Control of astrocytic Ca2+ signaling by nitric oxide-dependent S-nitrosylation of Ca2+ homeostasis modulator 1 channels.Biol Res. 2024 Apr 30;57(1):19. doi: 10.1186/s40659-024-00503-3. Biol Res. 2024. PMID: 38689353 Free PMC article.
-
S-acylation of Ca2+ transport proteins: molecular basis and functional consequences.Biochem Soc Trans. 2024 Feb 28;52(1):407-421. doi: 10.1042/BST20230818. Biochem Soc Trans. 2024. PMID: 38348884 Free PMC article. Review.
-
Intramolecular Disulfide Bonds for Biogenesis of CALHM1 Ion Channel Are Dispensable for Voltage-Dependent Activation.Mol Cells. 2021 Oct 31;44(10):758-769. doi: 10.14348/molcells.2021.0131. Mol Cells. 2021. PMID: 34711692 Free PMC article.
-
CALHM1/CALHM3 channel is intrinsically sorted to the basolateral membrane of epithelial cells including taste cells.Sci Rep. 2019 Feb 25;9(1):2681. doi: 10.1038/s41598-019-39593-5. Sci Rep. 2019. PMID: 30804437 Free PMC article.
-
ATP Release Channels.Int J Mol Sci. 2018 Mar 11;19(3):808. doi: 10.3390/ijms19030808. Int J Mol Sci. 2018. PMID: 29534490 Free PMC article. Review.
References
-
- Desgranges B. et al.. The neural substrates of episodic memory impairment in Alzheimer’s disease as revealed by FDG-PET: relationship to degree of deterioration. Brain 125, 1116–1124 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

