Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity
- PMID: 27066838
- PMCID: PMC4828653
- DOI: 10.1038/srep24242
Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity
Abstract
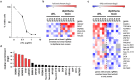
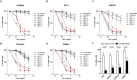
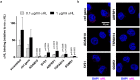
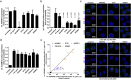
Staphylococcus aureus causes a wide variety of infections and antibiotic resistant strains are a major problem in hospitals. One of the best studied virulence factors of S. aureus is the pore-forming toxin alpha hemolysin (αHL) whose mechanism of action is incompletely understood. We performed a genome-wide loss-of-function screen using CRISPR/Cas9 technology to identify host targets required for αHL susceptibility in human myeloid cells. We found gRNAs for ten genes enriched after intoxication with αHL and focused on the top five hits. Besides a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), the host receptor for αHL, we identified three proteins, Sys1 golgi trafficking protein (SYS1), ADP-ribosylation factor 1 (ARFRP1), and tetraspanin-14 (TSPAN14) which regulate the presentation of ADAM10 on the plasma membrane post-translationally. Interestingly, we also showed that cells lacking sphingomyelin synthase 1 (SGMS1) resist αHL intoxication, but have only a slightly reduced ADAM10 surface expression. SGMS1 regulates lipid raft formation, suggesting that αHL requires these membrane microdomains for attachment and cytotoxicity.
Figures
Similar articles
-
Dissecting the role of ADAM10 as a mediator of Staphylococcus aureus α-toxin action.Biochem J. 2016 Jul 1;473(13):1929-40. doi: 10.1042/BCJ20160062. Epub 2016 May 4. Biochem J. 2016. PMID: 27147619
-
Staphylococcus aureus α-Toxin's Close Contacts Ensure the Kill.Trends Microbiol. 2019 Feb;27(2):89-90. doi: 10.1016/j.tim.2018.11.010. Epub 2018 Dec 13. Trends Microbiol. 2019. PMID: 30554769
-
The adherens junctions control susceptibility to Staphylococcus aureus α-toxin.Proc Natl Acad Sci U S A. 2015 Nov 17;112(46):14337-42. doi: 10.1073/pnas.1510265112. Epub 2015 Oct 21. Proc Natl Acad Sci U S A. 2015. PMID: 26489655 Free PMC article.
-
Staphylococcus aureus α-toxin: nearly a century of intrigue.Toxins (Basel). 2013 Jun;5(6):1140-66. doi: 10.3390/toxins5061140. Toxins (Basel). 2013. PMID: 23888516 Free PMC article. Review.
-
The keratinocyte as a target for staphylococcal bacterial toxins.J Investig Dermatol Symp Proc. 2001 Dec;6(3):225-30. doi: 10.1046/j.0022-202x.2001.00045.x. J Investig Dermatol Symp Proc. 2001. PMID: 11924832 Review.
Cited by
-
Sphingomyelin Depletion from Plasma Membranes of Human Airway Epithelial Cells Completely Abrogates the Deleterious Actions of S. aureus Alpha-Toxin.Toxins (Basel). 2019 Feb 20;11(2):126. doi: 10.3390/toxins11020126. Toxins (Basel). 2019. PMID: 30791542 Free PMC article.
-
Identification of SYS1 as a Host Factor Required for Shiga Toxin-Mediated Cytotoxicity in Vero Cells.Int J Mol Sci. 2021 May 6;22(9):4936. doi: 10.3390/ijms22094936. Int J Mol Sci. 2021. PMID: 34066520 Free PMC article.
-
Exploring the modulatory impact of isosakuranetin on Staphylococcus aureus: Inhibition of sortase A activity and α-haemolysin expression.Virulence. 2023 Dec;14(1):2260675. doi: 10.1080/21505594.2023.2260675. Epub 2023 Sep 28. Virulence. 2023. PMID: 37733916 Free PMC article.
-
The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex.J Biol Chem. 2020 Sep 4;295(36):12822-12839. doi: 10.1074/jbc.RA120.012601. Epub 2020 Feb 28. J Biol Chem. 2020. PMID: 32111735 Free PMC article.
-
Intermedilysin cytolytic activity depends on heparan sulfates and membrane composition.PLoS Genet. 2021 Feb 12;17(2):e1009387. doi: 10.1371/journal.pgen.1009387. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33577603 Free PMC article.
References
-
- Spaan A. N., Surewaard B. G. J., Nijland R. & van Strijp J. A. G. Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu. Rev. Microbiol. 67, 629–50 (2013). - PubMed
-
- Bartlett A. H., Foster T. J., Hayashida A. & Park P. W. Alpha-toxin facilitates the generation of CXC chemokine gradients and stimulates neutrophil homing in Staphylococcus aureus pneumonia. J. Infect. Dis. 198, 1529–1535 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

