Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design
- PMID: 27053554
- PMCID: PMC4907239
- DOI: 10.1128/JVI.03246-15
Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design
Abstract
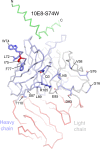
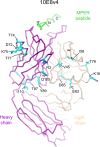
Extraordinary antibodies capable of near pan-neutralization of HIV-1 have been identified. One of the broadest is antibody 10E8, which recognizes the membrane-proximal external region (MPER) of the HIV-1 envelope and neutralizes >95% of circulating HIV-1 strains. If delivered passively, 10E8 might serve to prevent or treat HIV-1 infection. Antibody 10E8, however, is markedly less soluble than other antibodies. Here, we describe the use of both structural biology and somatic variation to develop optimized versions of 10E8 with increased solubility. From the structure of 10E8, we identified a prominent hydrophobic patch; reversion of four hydrophobic residues in this patch to their hydrophilic germ line counterparts resulted in an ∼10-fold decrease in turbidity. We also used somatic variants of 10E8, identified previously by next-generation sequencing, to optimize heavy and light chains; this process yielded several improved variants. Of these, variant 10E8v4 with 26 changes versus the parent 10E8 was the most soluble, with a paratope we showed crystallographically to be virtually identical to that of 10E8, a potency on a panel of 200 HIV-1 isolates also similar to that of 10E8, and a half-life in rhesus macaques of ∼10 days. An anomaly in 10E8v4 size exclusion chromatography that appeared to be related to conformational isomerization was resolved by engineering an interchain disulfide. Thus, by combining a structure-based approach with natural variation in potency and solubility from the 10E8 lineage, we successfully created variants of 10E8 which retained the potency and extraordinary neutralization breadth of the parent 10E8 but with substantially increased solubility.
Importance: Antibody 10E8 could be used to prevent HIV-1 infection, if manufactured and delivered economically. It suffers, however, from issues of solubility, which impede manufacturing. We hypothesized that the physical characteristic of 10E8 could be improved through rational design, without compromising breadth and potency. We used structural biology to identify hydrophobic patches on 10E8, which did not appear to be involved in 10E8 function. Reversion of hydrophobic residues in these patches to their hydrophilic germ line counterparts increased solubility. Next, clues from somatic variants of 10E8, identified by next-generation sequencing, were incorporated. A combination of structure-based design and somatic variant optimization led to 10E8v4, with substantially improved solubility and similar potency compared to the parent 10E8. The cocrystal structure of antibody 10E8v4 with its HIV-1 epitope was highly similar to that with the parent 10E8, despite 26 alterations in sequence and substantially improved solubility. Antibody 10E8v4 may be suitable for manufacturing.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.PLoS One. 2016 Jun 14;11(6):e0157409. doi: 10.1371/journal.pone.0157409. eCollection 2016. PLoS One. 2016. PMID: 27299673 Free PMC article.
-
Functional Optimization of Broadly Neutralizing HIV-1 Antibody 10E8 by Promotion of Membrane Interactions.J Virol. 2018 Mar 28;92(8):e02249-17. doi: 10.1128/JVI.02249-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386285 Free PMC article.
-
Surface-Matrix Screening Identifies Semi-specific Interactions that Improve Potency of a Near Pan-reactive HIV-1-Neutralizing Antibody.Cell Rep. 2018 Feb 13;22(7):1798-1809. doi: 10.1016/j.celrep.2018.01.023. Cell Rep. 2018. PMID: 29444432 Free PMC article.
-
Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization.Curr Opin HIV AIDS. 2017 May;12(3):250-256. doi: 10.1097/COH.0000000000000364. Curr Opin HIV AIDS. 2017. PMID: 28422789 Review.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
Cited by
-
Optimization of therapeutic antibodies.Antib Ther. 2021 Feb 18;4(1):45-54. doi: 10.1093/abt/tbab003. eCollection 2021 Jan. Antib Ther. 2021. PMID: 33928235 Free PMC article. Review.
-
Antibody Light Chains: Key to Increased Monoclonal Antibody Yields in Expi293 Cells?Antibodies (Basel). 2022 May 18;11(2):37. doi: 10.3390/antib11020037. Antibodies (Basel). 2022. PMID: 35645210 Free PMC article.
-
Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.iScience. 2021 Aug 17;24(9):102987. doi: 10.1016/j.isci.2021.102987. eCollection 2021 Sep 24. iScience. 2021. PMID: 34505005 Free PMC article.
-
Peripheral Membrane Interactions Boost the Engagement by an Anti-HIV-1 Broadly Neutralizing Antibody.J Biol Chem. 2017 Mar 31;292(13):5571-5583. doi: 10.1074/jbc.M117.775429. Epub 2017 Feb 17. J Biol Chem. 2017. PMID: 28213514 Free PMC article.
-
Enhanced Potency of a Broad H7N9-Neutralizing Antibody HNIgGA6 Through Structure-Based Design.Front Microbiol. 2020 Jun 19;11:1313. doi: 10.3389/fmicb.2020.01313. eCollection 2020. Front Microbiol. 2020. PMID: 32636820 Free PMC article.
References
-
- Corti D, Langedijk JPM, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. doi:10.1371/journal.pone.0008805. - DOI - PMC - PubMed
-
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang YP, Zhang ZH, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Karim SSA, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi:10.1038/nature13036. - DOI - PMC - PubMed
-
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, Mcbride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi:10.1016/j.immuni.2014.04.009. - DOI - PMC - PubMed
-
- Huang JH, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi:10.1038/nature13601. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

