X-ray structures and mechanism of the human serotonin transporter
- PMID: 27049939
- PMCID: PMC4898786
- DOI: 10.1038/nature17629
X-ray structures and mechanism of the human serotonin transporter
Abstract
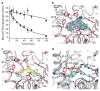
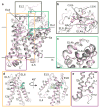
The serotonin transporter (SERT) terminates serotonergic signalling through the sodium- and chloride-dependent reuptake of neurotransmitter into presynaptic neurons. SERT is a target for antidepressant and psychostimulant drugs, which block reuptake and prolong neurotransmitter signalling. Here we report X-ray crystallographic structures of human SERT at 3.15 Å resolution bound to the antidepressants (S)-citalopram or paroxetine. Antidepressants lock SERT in an outward-open conformation by lodging in the central binding site, located between transmembrane helices 1, 3, 6, 8 and 10, directly blocking serotonin binding. We further identify the location of an allosteric site in the complex as residing at the periphery of the extracellular vestibule, interposed between extracellular loops 4 and 6 and transmembrane helices 1, 6, 10 and 11. Occupancy of the allosteric site sterically hinders ligand unbinding from the central site, providing an explanation for the action of (S)-citalopram as an allosteric ligand. These structures define the mechanism of antidepressant action in SERT, and provide blueprints for future drug design.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures




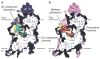
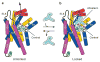
Comment in
-
Structural biology: Antidepressants at work.Nature. 2016 Apr 21;532(7599):320-1. doi: 10.1038/nature17883. Epub 2016 Apr 6. Nature. 2016. PMID: 27049942 No abstract available.
Similar articles
-
The mechanism of a high-affinity allosteric inhibitor of the serotonin transporter.Nat Commun. 2020 Mar 20;11(1):1491. doi: 10.1038/s41467-020-15292-y. Nat Commun. 2020. PMID: 32198394 Free PMC article.
-
Identification of an allosteric modulator of the serotonin transporter with novel mechanism of action.Neuropharmacology. 2013 Sep;72:282-90. doi: 10.1016/j.neuropharm.2013.04.026. Epub 2013 Apr 28. Neuropharmacology. 2013. PMID: 23632081
-
Steric hindrance mutagenesis in the conserved extracellular vestibule impedes allosteric binding of antidepressants to the serotonin transporter.J Biol Chem. 2012 Nov 16;287(47):39316-26. doi: 10.1074/jbc.M112.371765. Epub 2012 Sep 24. J Biol Chem. 2012. PMID: 23007398 Free PMC article.
-
Y95 and E444 interaction required for high-affinity S-citalopram binding in the human serotonin transporter.ACS Chem Neurosci. 2011 Feb 16;2(2):75-81. doi: 10.1021/cn100066p. Epub 2010 Oct 27. ACS Chem Neurosci. 2011. PMID: 22778858 Free PMC article. Review.
-
Escitalopram, an antidepressant with an allosteric effect at the serotonin transporter--a review of current understanding of its mechanism of action.Psychopharmacology (Berl). 2012 Jan;219(1):1-13. doi: 10.1007/s00213-011-2463-5. Epub 2011 Sep 8. Psychopharmacology (Berl). 2012. PMID: 21901317 Review.
Cited by
-
Fluorescent non-canonical amino acid provides insight into the human serotonin transporter.Nat Commun. 2024 Oct 27;15(1):9267. doi: 10.1038/s41467-024-53584-9. Nat Commun. 2024. PMID: 39463388 Free PMC article.
-
In silico structural studies on the vesicular neutral amino acid transporter NTT4 (SLC6A17).Comput Struct Biotechnol J. 2024 Sep 10;23:3342-3347. doi: 10.1016/j.csbj.2024.09.004. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39310277 Free PMC article.
-
Alternating access of a bacterial homolog of neurotransmitter: sodium symporters determined from AlphaFold2 ensembles and DEER spectroscopy.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2406063121. doi: 10.1073/pnas.2406063121. Epub 2024 Sep 20. Proc Natl Acad Sci U S A. 2024. PMID: 39302996
-
Substrate binding and inhibition mechanism of norepinephrine transporter.Nature. 2024 Sep;633(8029):473-479. doi: 10.1038/s41586-024-07810-5. Epub 2024 Aug 14. Nature. 2024. PMID: 39143211
-
Structure of the human dopamine transporter and mechanisms of inhibition.Nature. 2024 Aug;632(8025):672-677. doi: 10.1038/s41586-024-07739-9. Epub 2024 Aug 7. Nature. 2024. PMID: 39112705 Free PMC article.
References
-
- Rapport MM, Green AA, Page IH. Serum vasoconstrictor, serotonin; isolation and characterization. J Biol Chem. 1948;176:1243–1251. - PubMed
-
- Blackburn KJ, French PC, Merrills RJ. 5-hydroxytryptamine uptake by rat brain in vitro. Life Sci. 1967;6:1653–1663. - PubMed
-
- Carlsson A, Fuxe K, Ungerstedt U. The effect of imipramine on central 5-hydroxytryptamine neurons. J Pharm Pharmacol. 1968;20:150–151. - PubMed
-
- Glowinski J, Axelrod J. Inhibition of uptake of tritiated-noradrenaline in the intact rat brain by imipramine and structurally related compounds. Nature. 1964;204:1318–1319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

