Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing
- PMID: 27043410
- PMCID: PMC4972405
- DOI: 10.1038/ni.3412
Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing
Abstract
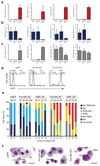
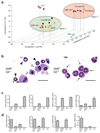
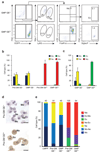
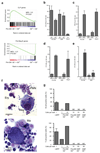
According to current models of hematopoiesis, lymphoid-primed multi-potent progenitors (LMPPs) (Lin(-)Sca-1(+)c-Kit(+)CD34(+)Flt3(hi)) and common myeloid progenitors (CMPs) (Lin(-)Sca-1(+)c-Kit(+)CD34(+)CD41(hi)) establish an early branch point for separate lineage-commitment pathways from hematopoietic stem cells, with the notable exception that both pathways are proposed to generate all myeloid innate immune cell types through the same myeloid-restricted pre-granulocyte-macrophage progenitor (pre-GM) (Lin(-)Sca-1(-)c-Kit(+)CD41(-)FcγRII/III(-)CD150(-)CD105(-)). By single-cell transcriptome profiling of pre-GMs, we identified distinct myeloid differentiation pathways: a pathway expressing the gene encoding the transcription factor GATA-1 generated mast cells, eosinophils, megakaryocytes and erythroid cells, and a pathway lacking expression of that gene generated monocytes, neutrophils and lymphocytes. These results identify an early hematopoietic-lineage bifurcation that separates the myeloid lineages before their segregation from other hematopoietic-lineage potential.
Conflict of interest statement
statement. The authors have no competing financial interests.
Figures

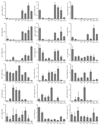
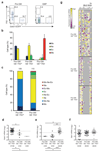
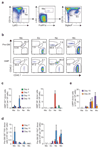
Comment in
-
Eosinophils and mast cells: a lineage apart.Nat Immunol. 2016 May 19;17(6):609-11. doi: 10.1038/ni.3446. Nat Immunol. 2016. PMID: 27196510 No abstract available.
Similar articles
-
CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor.Stem Cells. 2015 Mar;33(3):976-87. doi: 10.1002/stem.1906. Stem Cells. 2015. PMID: 25446279
-
Identification of Flt3⁺CD150⁻ myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential.Blood. 2011 Sep 8;118(10):2723-32. doi: 10.1182/blood-2010-09-309989. Epub 2011 Jul 26. Blood. 2011. PMID: 21791413 Free PMC article.
-
Combined signaling through interleukin-7 receptors and flt3 but not c-kit potently and selectively promotes B-cell commitment and differentiation from uncommitted murine bone marrow progenitor cells.Blood. 1996 Aug 15;88(4):1256-65. Blood. 1996. PMID: 8695843
-
Biological and molecular evidence for existence of lymphoid-primed multipotent progenitors.Ann N Y Acad Sci. 2007 Jun;1106:89-94. doi: 10.1196/annals.1392.023. Epub 2007 Apr 18. Ann N Y Acad Sci. 2007. PMID: 17442777 Review.
-
Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells.Leukemia. 1996 Apr;10(4):588-99. Leukemia. 1996. PMID: 8618433 Review.
Cited by
-
CD34 and EPCR coordinately enrich functional murine hematopoietic stem cells under normal and inflammatory conditions.Exp Hematol. 2020 Jan;81:1-15.e6. doi: 10.1016/j.exphem.2019.12.003. Epub 2019 Dec 18. Exp Hematol. 2020. PMID: 31863798 Free PMC article.
-
Allogeneic transplantation of peripheral blood stem cell grafts results in a massive decrease of primitive hematopoietic progenitor frequencies in reconstituted bone marrows.Bone Marrow Transplant. 2020 Jan;55(1):100-109. doi: 10.1038/s41409-019-0645-7. Epub 2019 Aug 21. Bone Marrow Transplant. 2020. PMID: 31435034
-
Kinetics of adult hematopoietic stem cell differentiation in vivo.J Exp Med. 2018 Nov 5;215(11):2815-2832. doi: 10.1084/jem.20180136. Epub 2018 Oct 5. J Exp Med. 2018. PMID: 30291161 Free PMC article.
-
Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints.Blood. 2019 Mar 7;133(10):1119-1129. doi: 10.1182/blood-2018-08-862292. Epub 2018 Dec 27. Blood. 2019. PMID: 30591526 Free PMC article.
-
Mast Cells and Their Progenitors in Allergic Asthma.Front Immunol. 2019 May 29;10:821. doi: 10.3389/fimmu.2019.00821. eCollection 2019. Front Immunol. 2019. PMID: 31191511 Free PMC article. Review.
References
-
- Koenderman L, Buurman W, Daha MR. The innate immune response. Immunol Lett. 2014;162(2PB):95–102. - PubMed
-
- Metcalf D. Lineage commitment and maturation in hematopoietic cells: the case for extrinsic regulation. Blood. 1998;92(2):345–347. discussion 352. - PubMed
-
- Sadrzadeh H, Abdel-Wahab O, Fathi AT. Molecular alterations underlying eosinophilic and mast cell malignancies. Discov Med. 2011;12(67):481–493. - PubMed
-
- Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12009/16/MRC_/Medical Research Council/United Kingdom
- MR/M00919X/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12025 /MRC_/Medical Research Council/United Kingdom
- 11-0724/AICR_/Worldwide Cancer Research/United Kingdom
- MC_UU_12009/5/MRC_/Medical Research Council/United Kingdom
- MR/L006340/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_12020/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G0501838/MRC_/Medical Research Council/United Kingdom
- 087371/WT_/Wellcome Trust/United Kingdom
- G0801073/MRC_/Medical Research Council/United Kingdom
- MR/K017047/1/MRC_/Medical Research Council/United Kingdom
- G0900892/MRC_/Medical Research Council/United Kingdom
- G0701761/MRC_/Medical Research Council/United Kingdom
- G0902418/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/7/MRC_/Medical Research Council/United Kingdom
- MR/L012766/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

