Brinp1(-/-) mice exhibit autism-like behaviour, altered memory, hyperactivity and increased parvalbumin-positive cortical interneuron density
- PMID: 27042284
- PMCID: PMC4818446
- DOI: 10.1186/s13229-016-0079-7
Brinp1(-/-) mice exhibit autism-like behaviour, altered memory, hyperactivity and increased parvalbumin-positive cortical interneuron density
Abstract
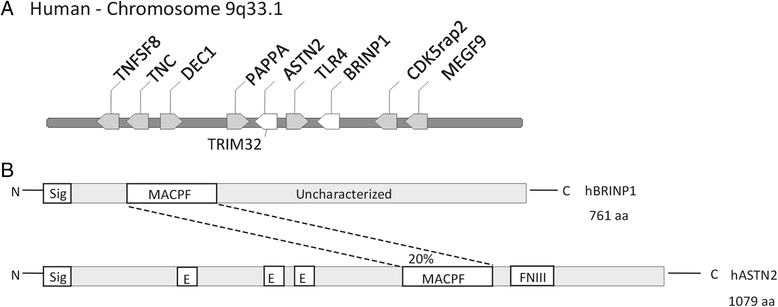
Background: BMP/RA-inducible neural-specific protein 1 (Brinp1) is highly conserved in vertebrates, and continuously expressed in the neocortex, hippocampus, olfactory bulb and cerebellum from mid-embryonic development through to adulthood.
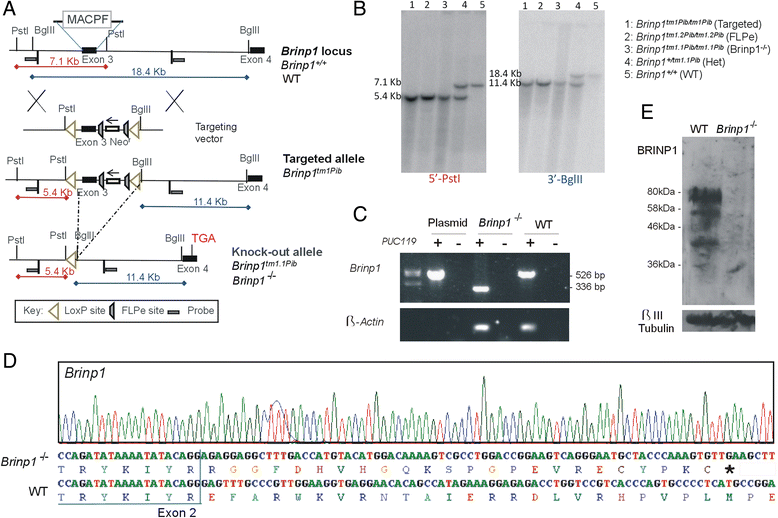
Methods: Brinp1 knock-out (Brinp1(-/-)) mice were generated by Cre-recombinase-mediated removal of the third exon of Brinp1. Knock-out mice were characterised by behavioural phenotyping, immunohistochemistry and expression analysis of the developing and adult brain.
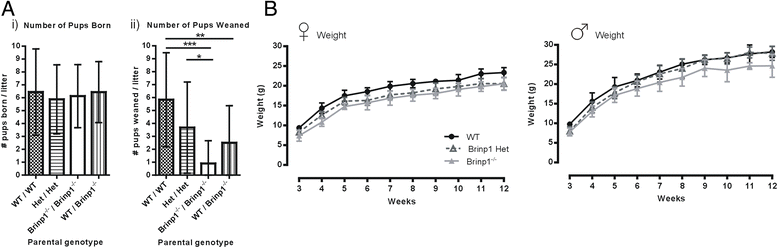
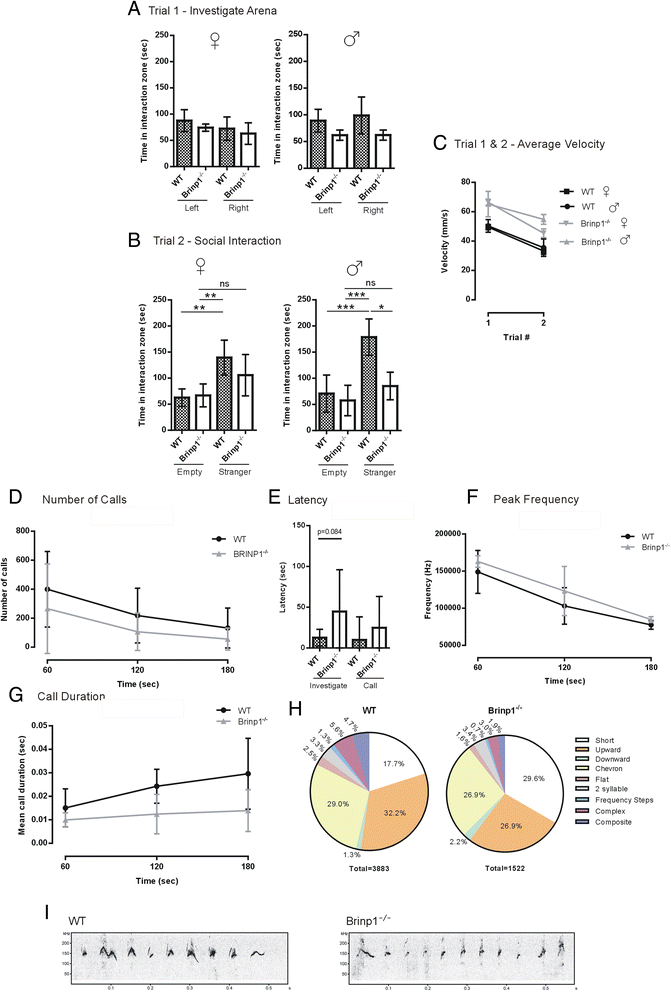
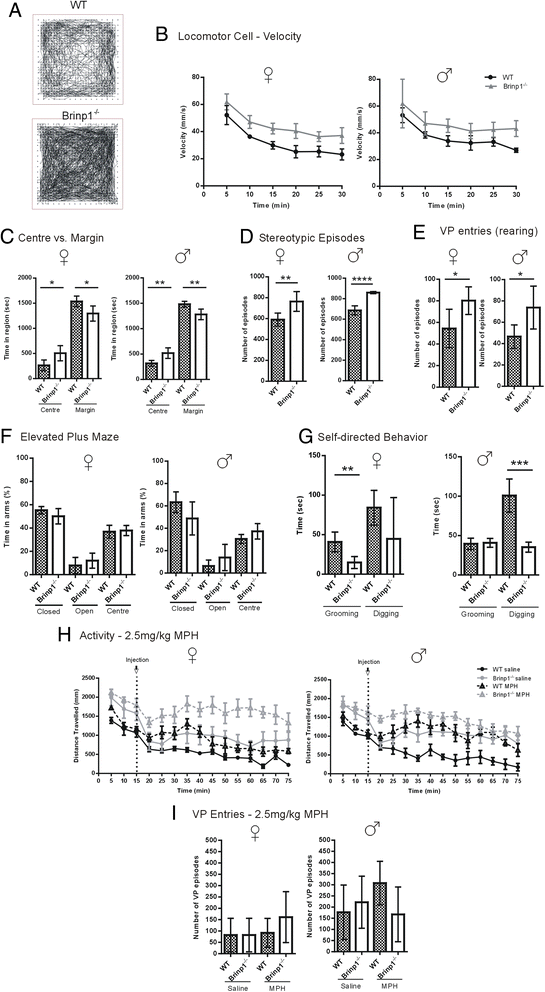
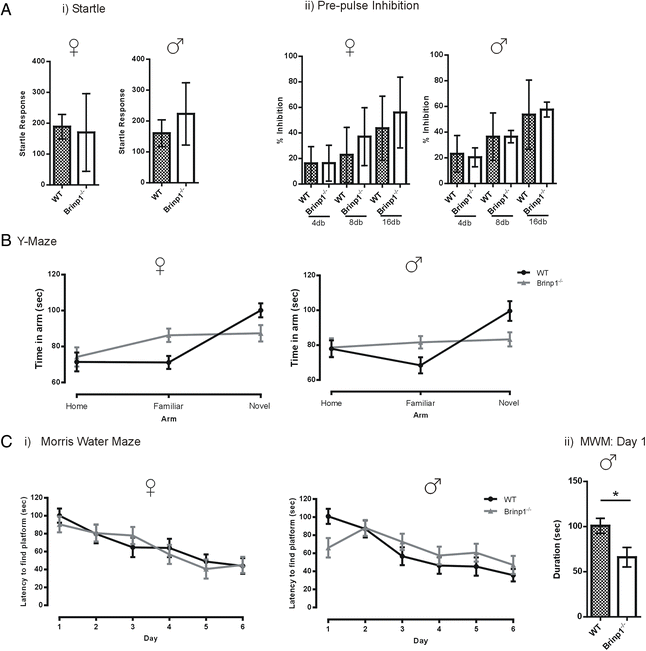
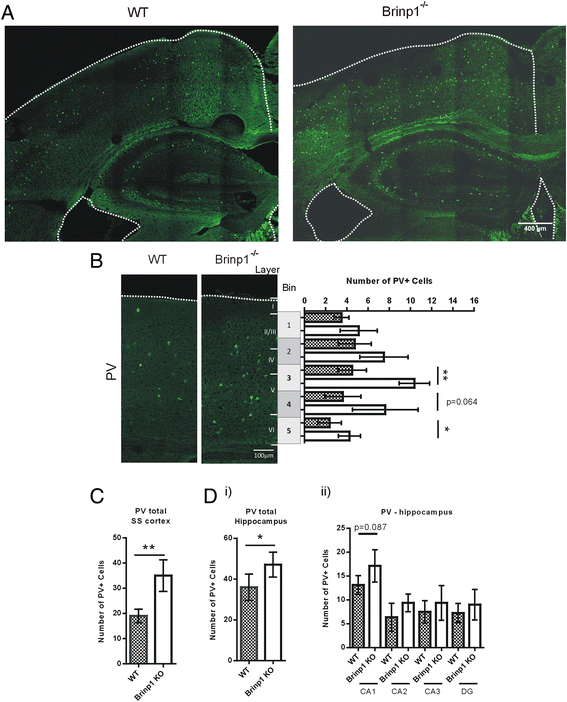
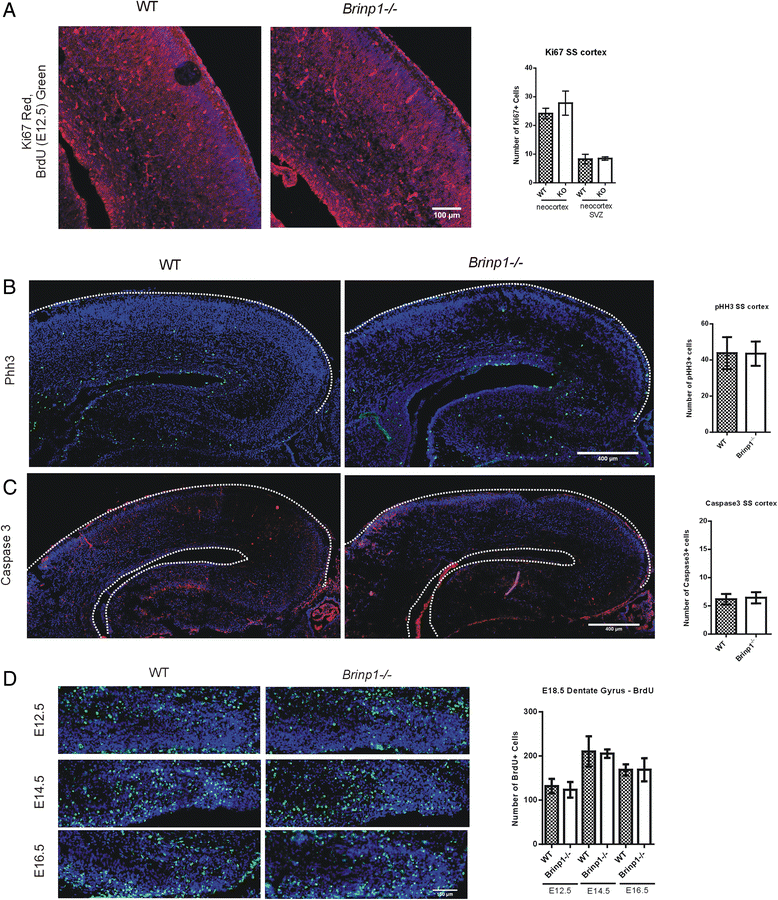
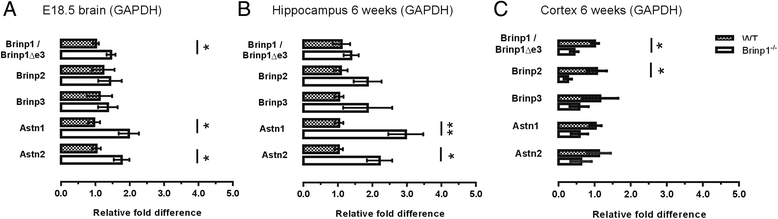
Results: Absence of Brinp1 during development results in a behavioural phenotype resembling autism spectrum disorder (ASD), in which knock-out mice show reduced sociability and changes in vocalisation capacity. In addition, Brinp1(-/-) mice exhibit hyper-locomotor activity, have impaired short-term memory, and exhibit poor reproductive success. Brinp1(-/-) mice show increased density of parvalbumin-expressing interneurons in the adult mouse brain. Brinp1(-/-) mice do not show signs of altered neural precursor proliferation or increased apoptosis during late embryonic brain development. The expression of the related neuronal migration genes Astn1 and Astn2 is increased in the brains of Brinp1(-/-) mice, suggesting that they may ameliorate the effects of Brinp1 loss.
Conclusions: Brinp1 plays an important role in normal brain development and function by influencing neuronal distribution within the cortex. The increased cortical PV-positive interneuron density and altered behaviour of Brinp1(-/-) mice resemble features of a subset of human neurological disorders; namely autism spectrum disorder (ASD) and the hyperactivity aspect of attention deficit hyperactivity disorder (ADHD).
Keywords: Autism spectrum disorder; Brinp1; Cortex; Hyperactivity; Interneuron; Knock-out; Neurodevelopment; Parvalbumin.
Figures
Similar articles
-
Absence of BRINP1 in mice causes increase of hippocampal neurogenesis and behavioral alterations relevant to human psychiatric disorders.Mol Brain. 2014 Feb 14;7:12. doi: 10.1186/1756-6606-7-12. Mol Brain. 2014. PMID: 24528488 Free PMC article.
-
Decreased parvalbumin and somatostatin neurons in medial prefrontal cortex in BRINP1-KO mice.Neurosci Lett. 2018 Sep 14;683:82-88. doi: 10.1016/j.neulet.2018.06.050. Epub 2018 Jun 28. Neurosci Lett. 2018. PMID: 29960053
-
Mice lacking Astn2 have ASD-like behaviors and altered cerebellar circuit properties.Proc Natl Acad Sci U S A. 2024 Aug 20;121(34):e2405901121. doi: 10.1073/pnas.2405901121. Epub 2024 Aug 16. Proc Natl Acad Sci U S A. 2024. PMID: 39150780 Free PMC article.
-
The Role of Parvalbumin Interneurons in Autism Spectrum Disorder.J Neurosci Res. 2024 Oct;102(10):e25391. doi: 10.1002/jnr.25391. J Neurosci Res. 2024. PMID: 39400385 Review.
-
Autism spectrum disorder: neuropathology and animal models.Acta Neuropathol. 2017 Oct;134(4):537-566. doi: 10.1007/s00401-017-1736-4. Epub 2017 Jun 5. Acta Neuropathol. 2017. PMID: 28584888 Free PMC article. Review.
Cited by
-
Maternal separation with early weaning impairs neuron-glia integrity: non-invasive evaluation and substructure demonstration.Sci Rep. 2020 Nov 10;10(1):19440. doi: 10.1038/s41598-020-76640-y. Sci Rep. 2020. PMID: 33173142 Free PMC article.
-
PlexinA1 deficiency in BALB/cAJ mice leads to excessive self-grooming and reduced prepulse inhibition.IBRO Rep. 2020 Oct 22;9:276-289. doi: 10.1016/j.ibror.2020.10.004. eCollection 2020 Dec. IBRO Rep. 2020. PMID: 33163687 Free PMC article.
-
Mice Lacking Brinp2 or Brinp3, or Both, Exhibit Behaviors Consistent with Neurodevelopmental Disorders.Front Behav Neurosci. 2016 Oct 25;10:196. doi: 10.3389/fnbeh.2016.00196. eCollection 2016. Front Behav Neurosci. 2016. PMID: 27826231 Free PMC article.
-
Foxp1 regulation of neonatal vocalizations via cortical development.Genes Dev. 2017 Oct 15;31(20):2039-2055. doi: 10.1101/gad.305037.117. Epub 2017 Nov 14. Genes Dev. 2017. PMID: 29138280 Free PMC article.
-
Genetic Variation in the ASTN2 Locus in Cardiovascular, Metabolic and Psychiatric Traits: Evidence for Pleiotropy Rather Than Shared Biology.Genes (Basel). 2021 Jul 31;12(8):1194. doi: 10.3390/genes12081194. Genes (Basel). 2021. PMID: 34440368 Free PMC article.
References
-
- Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J Clin Psychiatry. 2005;66(Suppl 10):3–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

