Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity
- PMID: 27040782
- PMCID: PMC4846481
- DOI: 10.1016/j.cub.2016.02.061
Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity
Abstract
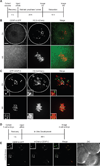
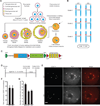
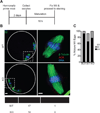
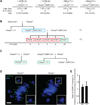
Centromeres control genetic inheritance by directing chromosome segregation but are not genetically encoded themselves. Rather, centromeres are defined by nucleosomes containing CENP-A, a histone H3 variant [1]. In cycling somatic cells, centromere identity is maintained by an established cell-cycle-coupled CENP-A chromatin assembly pathway, but how centromeres are inherited through the mammalian female germline is unclear because of the long (months to decades) prophase I arrest. Here we show that mouse oocytes retain the pool of CENP-A nucleosomes assembled before birth, and that this pool is sufficient for centromere function, fertility, and genome transmission to embryos. Indeed, oocytes lack any measurable CENP-A nucleosome assembly through the entire fertile lifespan of the female (>1 year). Thus, the remarkable stability of CENP-A nucleosomes confers transgenerational centromere identity in mammals.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Oogenesis: Ageing Oocyte Chromosomes Rely on Amazing Protein Stability.Curr Biol. 2016 Apr 25;26(8):R329-31. doi: 10.1016/j.cub.2016.02.059. Curr Biol. 2016. PMID: 27115691
Similar articles
-
Centromere-specifying nucleosomes persist in aging mouse oocytes in the absence of nascent assembly.Curr Biol. 2023 Sep 11;33(17):3759-3765.e3. doi: 10.1016/j.cub.2023.07.032. Epub 2023 Aug 14. Curr Biol. 2023. PMID: 37582374 Free PMC article.
-
Maternal inheritance of centromeres through the germline.Curr Top Dev Biol. 2020;140:35-54. doi: 10.1016/bs.ctdb.2020.03.004. Epub 2020 Apr 25. Curr Top Dev Biol. 2020. PMID: 32591081 Review.
-
Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential.Dev Cell. 2019 Oct 7;51(1):35-48.e7. doi: 10.1016/j.devcel.2019.07.016. Epub 2019 Aug 15. Dev Cell. 2019. PMID: 31422918 Free PMC article.
-
Centromere-specifying nucleosomes persist in aging mouse oocytes in the absence of nascent assembly.bioRxiv [Preprint]. 2023 May 18:2023.05.18.541332. doi: 10.1101/2023.05.18.541332. bioRxiv. 2023. Update in: Curr Biol. 2023 Sep 11;33(17):3759-3765.e3. doi: 10.1016/j.cub.2023.07.032 PMID: 37292821 Free PMC article. Updated. Preprint.
-
Orchestrating the Specific Assembly of Centromeric Nucleosomes.Prog Mol Subcell Biol. 2017;56:165-192. doi: 10.1007/978-3-319-58592-5_7. Prog Mol Subcell Biol. 2017. PMID: 28840237 Free PMC article. Review.
Cited by
-
The Mis6 inner kinetochore subcomplex maintains CENP-A nucleosomes against centromeric non-coding transcription during mitosis.Commun Biol. 2022 Aug 15;5(1):818. doi: 10.1038/s42003-022-03786-y. Commun Biol. 2022. PMID: 35970865 Free PMC article.
-
Asymmetric assembly of centromeres epigenetically regulates stem cell fate.J Cell Biol. 2020 Apr 6;219(4):e201910084. doi: 10.1083/jcb.201910084. J Cell Biol. 2020. PMID: 32328637 Free PMC article.
-
Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8.Nature. 2021 Mar;591(7851):671-676. doi: 10.1038/s41586-021-03279-8. Epub 2021 Mar 3. Nature. 2021. PMID: 33658710 Free PMC article.
-
Centromere DNA Destabilizes H3 Nucleosomes to Promote CENP-A Deposition during the Cell Cycle.Curr Biol. 2018 Dec 17;28(24):3924-3936.e4. doi: 10.1016/j.cub.2018.10.049. Epub 2018 Nov 29. Curr Biol. 2018. PMID: 30503616 Free PMC article.
-
Auxin-inducible protein degradation as a novel approach for protein depletion and reverse genetic discoveries in mammalian oocytes†.Biol Reprod. 2019 Oct 25;101(4):704-718. doi: 10.1093/biolre/ioz113. Biol Reprod. 2019. PMID: 31299080 Free PMC article.
References
-
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 2005;7:1248–1255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

