Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses
- PMID: 27036003
- PMCID: PMC4843436
- DOI: 10.1073/pnas.1603106113
Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses
Abstract
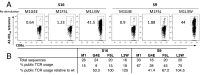
Memory CD8(+)T lymphocytes (CTLs) specific for antigenic peptides derived from internal viral proteins confer broad protection against distinct strains of influenza A virus (IAV). However, immune efficacy can be undermined by the emergence of escape mutants. To determine how T-cell receptor (TCR) composition relates to IAV epitope variability, we used ex vivo peptide-HLA tetramer enrichment and single-cell multiplex analysis to compare TCRs targeted to the largely conserved HLA-A*0201-M158and the hypervariable HLA-B*3501-NP418antigens. The TCRαβs for HLA-B*3501-NP418 (+)CTLs varied among individuals and across IAV strains, indicating that a range of mutated peptides will prime different NP418-specific CTL sets. Conversely, a dominant public TRAV27/TRBV19(+)TCRαβ was selected in HLA-A*0201(+)donors responding to M158 This public TCR cross-recognized naturally occurring M158variants complexed with HLA-A*0201. Ternary structures showed that induced-fit molecular mimicry underpins TRAV27/TRBV19(+)TCR specificity for the WT and mutant M158peptides, suggesting the possibility of universal CTL immunity in HLA-A*0201(+)individuals. Combined with the high population frequency of HLA-A*0201, these data potentially explain the relative conservation of M158 Moreover, our results suggest that vaccination strategies aimed at generating broad protection should incorporate variant peptides to elicit cross-reactive responses against other specificities, especially those that may be relatively infrequent among IAV-primed memory CTLs.
Keywords: T-cell receptor; human CD8+ T cells; influenza infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

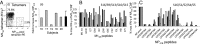





Similar articles
-
HLA-B*27:05 alters immunodominance hierarchy of universal influenza-specific CD8+ T cells.PLoS Pathog. 2020 Aug 4;16(8):e1008714. doi: 10.1371/journal.ppat.1008714. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32750095 Free PMC article.
-
Severity of Acute Infectious Mononucleosis Correlates with Cross-Reactive Influenza CD8 T-Cell Receptor Repertoires.mBio. 2017 Dec 5;8(6):e01841-17. doi: 10.1128/mBio.01841-17. mBio. 2017. PMID: 29208744 Free PMC article.
-
Variation at Extra-epitopic Amino Acid Residues Influences Suppression of Influenza Virus Replication by M158-66 Epitope-Specific CD8+ T Lymphocytes.J Virol. 2018 May 14;92(11):e00232-18. doi: 10.1128/JVI.00232-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29593036 Free PMC article.
-
[The impact of conservative and hypervariable immunodominant epitopes in internal proteins of the influenza A virus on cytotoxic T-cell immune responses].Vopr Virusol. 2015;60(1):11-6. Vopr Virusol. 2015. PMID: 26021066 Review. Russian.
-
T cell mediated immunity to influenza: mechanisms of viral control.Trends Immunol. 2014 Aug;35(8):396-402. doi: 10.1016/j.it.2014.06.004. Epub 2014 Jul 16. Trends Immunol. 2014. PMID: 25043801 Review.
Cited by
-
Challenging immunodominance of influenza-specific CD8+ T cell responses restricted by the risk-associated HLA-A*68:01 allomorph.Nat Commun. 2019 Dec 6;10(1):5579. doi: 10.1038/s41467-019-13346-4. Nat Commun. 2019. PMID: 31811120 Free PMC article.
-
Extracellular Vesicles and Their Use as Vehicles of Immunogens.Methods Mol Biol. 2022;2504:177-198. doi: 10.1007/978-1-0716-2341-1_13. Methods Mol Biol. 2022. PMID: 35467287
-
Prognostic factors and outcomes of COVID-19 cases in Ethiopia: multi-center cohort study protocol.BMC Infect Dis. 2021 Sep 16;21(1):956. doi: 10.1186/s12879-021-06652-0. BMC Infect Dis. 2021. PMID: 34530744 Free PMC article.
-
The presentation of SARS-CoV-2 peptides by the common HLA-A∗02:01 molecule.iScience. 2021 Feb 19;24(2):102096. doi: 10.1016/j.isci.2021.102096. Epub 2021 Jan 22. iScience. 2021. PMID: 33521593 Free PMC article.
-
Phenotypic and functional characterization of pharmacologically expanded Vγ9Vδ2 T cells in pigtail macaques.iScience. 2023 Feb 25;26(3):106269. doi: 10.1016/j.isci.2023.106269. eCollection 2023 Mar 17. iScience. 2023. PMID: 36936791 Free PMC article.
References
-
- Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19(10):1305–1312. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

