CCDC141 Mutation Identified in Anosmic Hypogonadotropic Hypogonadism (Kallmann Syndrome) Alters GnRH Neuronal Migration
- PMID: 27014940
- PMCID: PMC4870868
- DOI: 10.1210/en.2015-1846
CCDC141 Mutation Identified in Anosmic Hypogonadotropic Hypogonadism (Kallmann Syndrome) Alters GnRH Neuronal Migration
Abstract
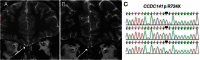
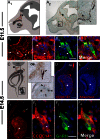
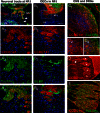
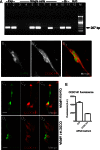
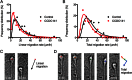
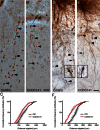
The first mutation in a gene associated with a neuronal migration disorder was identified in patients with Kallmann Syndrome, characterized by hypogonadotropic hypogonadism and anosmia. This pathophysiological association results from a defect in the development of the GnRH and the olfactory system. A recent genetic screening of Kallmann Syndrome patients revealed a novel mutation in CCDC141. Little is known about CCDC141, which encodes a coiled-coil domain containing protein. Here, we show that Ccdc141 is expressed in GnRH neurons and olfactory fibers and that knockdown of Ccdc141 reduces GnRH neuronal migration. Our findings in human patients and mouse models predict that CCDC141 takes part in embryonic migration of GnRH neurons enabling them to form a hypothalamic neuronal network to initiate pulsatile GnRH secretion and reproductive function.
Figures
Similar articles
-
CCDC141 Mutations in Idiopathic Hypogonadotropic Hypogonadism.J Clin Endocrinol Metab. 2017 Jun 1;102(6):1816-1825. doi: 10.1210/jc.2016-3391. J Clin Endocrinol Metab. 2017. PMID: 28324054 Free PMC article.
-
[GnRH deficiency: new insights from genetics].J Soc Biol. 2004;198(1):80-7. J Soc Biol. 2004. PMID: 15146960 Review. French.
-
Forebrain gonadotropin-releasing hormone neuronal development: insights from transgenic medaka and the relevance to X-linked Kallmann syndrome.Endocrinology. 2006 Mar;147(3):1076-84. doi: 10.1210/en.2005-0468. Epub 2005 Nov 17. Endocrinology. 2006. PMID: 16293668
-
Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism.Proc Natl Acad Sci U S A. 2007 Oct 30;104(44):17447-52. doi: 10.1073/pnas.0707173104. Epub 2007 Oct 24. Proc Natl Acad Sci U S A. 2007. PMID: 17959774 Free PMC article.
-
[Clinical and molecular aspects of congenital isolated hypogonadotropic hypogonadism].Arq Bras Endocrinol Metabol. 2011 Nov;55(8):501-11. doi: 10.1590/s0004-27302011000800002. Arq Bras Endocrinol Metabol. 2011. PMID: 22218430 Review. Portuguese.
Cited by
-
Genome-Wide Association Study of Accessory Atrioventricular Pathways.JAMA Cardiol. 2024 Nov 1;9(11):1053-1058. doi: 10.1001/jamacardio.2024.2684. JAMA Cardiol. 2024. PMID: 39230897
-
Increased Burden of Rare Sequence Variants in GnRH-Associated Genes in Women With Hypothalamic Amenorrhea.J Clin Endocrinol Metab. 2021 Mar 8;106(3):e1441-e1452. doi: 10.1210/clinem/dgaa609. J Clin Endocrinol Metab. 2021. PMID: 32870266 Free PMC article.
-
New findings in oligogenic inheritance of congenital hypogonadotropic hypogonadism.Arch Med Sci. 2020 Sep 18;18(2):353-364. doi: 10.5114/aoms.2020.98909. eCollection 2022. Arch Med Sci. 2020. PMID: 35316923 Free PMC article.
-
Exome sequencing of families from Ghana reveals known and candidate hearing impairment genes.Commun Biol. 2022 Apr 19;5(1):369. doi: 10.1038/s42003-022-03326-8. Commun Biol. 2022. PMID: 35440622 Free PMC article.
-
Whole exome sequencing and trio analysis to broaden the variant spectrum of genes in idiopathic hypogonadotropic hypogonadism.Asian J Androl. 2021 May-Jun;23(3):288-293. doi: 10.4103/aja.aja_65_20. Asian J Androl. 2021. PMID: 33208564 Free PMC article.
References
-
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. - PubMed
-
- Franco B, Guioli S, Pragliola A, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. - PubMed
-
- Ballabio A, Camerino G. The gene for X-linked Kallmann syndrome: a human neuronal migration defect. Curr Opin Genet Dev. 1992;2:417–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

