Comparative genomic analysis of pre-epidemic and epidemic Zika virus strains for virological factors potentially associated with the rapidly expanding epidemic
- PMID: 26980239
- PMCID: PMC4820678
- DOI: 10.1038/emi.2016.48
Comparative genomic analysis of pre-epidemic and epidemic Zika virus strains for virological factors potentially associated with the rapidly expanding epidemic
Abstract
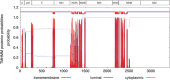
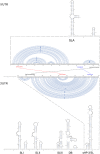
Less than 20 sporadic cases of human Zika virus (ZIKV) infection were reported in Africa and Asia before 2007, but large outbreaks involving up to 73% of the populations on the Pacific islands have started since 2007, and spread to the Americas in 2014. Moreover, the clinical manifestation of ZIKV infection has apparently changed, as evident by increasing reports of neurological complications, such as Guillain-Barré syndrome in adults and congenital anomalies in neonates. We comprehensively compared the genome sequences of pre-epidemic and epidemic ZIKV strains with complete genome or complete polyprotein sequences available in GenBank. Besides the reported phylogenetic clustering of the epidemic strains with the Asian lineage, we found that the topology of phylogenetic tree of all coding regions is the same except that of the non-structural 2B (NS2B) coding region. This finding was confirmed by bootscan analysis and multiple sequence alignment, which suggested the presence of a fragment of genetic recombination at NS2B with that of Spondweni virus. Moreover, the representative epidemic strain possesses one large bulge of nine bases instead of an external loop on the first stem-loop structure at the 3'-untranslated region just distal to the stop codon of the NS5 in the 1947 pre-epidemic prototype strain. Fifteen amino acid substitutions are found in the epidemic strains when compared with the pre-epidemic strains. As mutations in other flaviviruses can be associated with changes in virulence, replication efficiency, antigenic epitopes and host tropism, further studies would be important to ascertain the biological significance of these genomic changes.
Figures




Similar articles
-
Comparative analysis of protein evolution in the genome of pre-epidemic and epidemic Zika virus.Infect Genet Evol. 2017 Jul;51:74-85. doi: 10.1016/j.meegid.2017.03.012. Epub 2017 Mar 14. Infect Genet Evol. 2017. PMID: 28315476
-
Role of mutational reversions and fitness restoration in Zika virus spread to the Americas.Nat Commun. 2021 Jan 26;12(1):595. doi: 10.1038/s41467-020-20747-3. Nat Commun. 2021. PMID: 33500409 Free PMC article.
-
Phylogenetic analysis revealed the central roles of two African countries in the evolution and worldwide spread of Zika virus.Virol Sin. 2016 Apr;31(2):118-30. doi: 10.1007/s12250-016-3774-9. Epub 2016 Apr 26. Virol Sin. 2016. PMID: 27129451 Free PMC article.
-
The evolution of Zika virus from Asia to the Americas.Nat Rev Microbiol. 2019 Mar;17(3):131-139. doi: 10.1038/s41579-018-0134-9. Nat Rev Microbiol. 2019. PMID: 30617340 Review.
-
The Asian Lineage of Zika Virus: Transmission and Evolution in Asia and the Americas.Virol Sin. 2019 Feb;34(1):1-8. doi: 10.1007/s12250-018-0078-2. Epub 2019 Jan 25. Virol Sin. 2019. PMID: 30684211 Free PMC article. Review.
Cited by
-
Nuclear localization of Zika virus NS5 contributes to suppression of type I interferon production and response.J Gen Virol. 2021 Mar;102(3):001376. doi: 10.1099/jgv.0.001376. Epub 2019 Dec 18. J Gen Virol. 2021. PMID: 31859616 Free PMC article.
-
The Nuclear Pore Complex: A Target for NS3 Protease of Dengue and Zika Viruses.Viruses. 2020 May 26;12(6):583. doi: 10.3390/v12060583. Viruses. 2020. PMID: 32466480 Free PMC article.
-
Inhibition of Zika Virus Replication by Silvestrol.Viruses. 2018 Mar 27;10(4):149. doi: 10.3390/v10040149. Viruses. 2018. PMID: 29584632 Free PMC article.
-
Recent Perspectives on Genome, Transmission, Clinical Manifestation, Diagnosis, Therapeutic Strategies, Vaccine Developments, and Challenges of Zika Virus Research.Front Microbiol. 2017 Sep 14;8:1761. doi: 10.3389/fmicb.2017.01761. eCollection 2017. Front Microbiol. 2017. PMID: 28959246 Free PMC article. Review.
-
Zika virus replication in the mosquito Culex quinquefasciatus in Brazil.Emerg Microbes Infect. 2017 Aug 9;6(8):e69. doi: 10.1038/emi.2017.59. Emerg Microbes Infect. 2017. PMID: 28790458 Free PMC article.
References
-
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 1952; 46: 509–520. - PubMed
-
- Duffy MR, Chen TH, Hancock WT et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 2009; 360: 2536–2543. - PubMed
-
- Tognarelli J, Ulloa S, Villagra E et al. A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch Virol 2016; 151: 665–668. - PubMed
-
- Roth A, Mercier A, Lepers C et al. Concurrent outbreaks of dengue, chikungunya and Zika virus infections - an unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012-2014. Euro Surveill 2014; 19: pii: 20929. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
