Rhodopsin targeted transcriptional silencing by DNA-binding
- PMID: 26974343
- PMCID: PMC4805542
- DOI: 10.7554/eLife.12242
Rhodopsin targeted transcriptional silencing by DNA-binding
Abstract
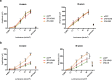
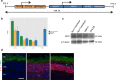
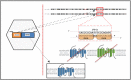
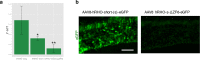
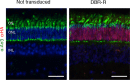
Transcription factors (TFs) operate by the combined activity of their DNA-binding domains (DBDs) and effector domains (EDs) enabling the coordination of gene expression on a genomic scale. Here we show that in vivo delivery of an engineered DNA-binding protein uncoupled from the repressor domain can produce efficient and gene-specific transcriptional silencing. To interfere with RHODOPSIN (RHO) gain-of-function mutations we engineered the ZF6-DNA-binding protein (ZF6-DB) that targets 20 base pairs (bp) of a RHOcis-regulatory element (CRE) and demonstrate Rho specific transcriptional silencing upon adeno-associated viral (AAV) vector-mediated expression in photoreceptors. The data show that the 20 bp-long genomic DNA sequence is necessary for RHO expression and that photoreceptor delivery of the corresponding cognate synthetic trans-acting factor ZF6-DB without the intrinsic transcriptional repression properties of the canonical ED blocks Rho expression with negligible genome-wide transcript perturbations. The data support DNA-binding-mediated silencing as a novel mode to treat gain-of-function mutations.
Keywords: AAV; gene expression; gene therapy; human biology; medicine; mouse; neurodegeneration; neuroscience; retina; rhodopsin; transcription; transcription factors; zinc finger.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
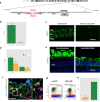

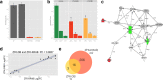
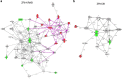
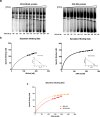
Similar articles
-
Challenging Safety and Efficacy of Retinal Gene Therapies by Retinogenesis.Int J Mol Sci. 2021 May 28;22(11):5767. doi: 10.3390/ijms22115767. Int J Mol Sci. 2021. PMID: 34071252 Free PMC article.
-
Targeting and silencing of rhodopsin by ectopic expression of the transcription factor KLF15.JCI Insight. 2017 Dec 21;2(24):e96560. doi: 10.1172/jci.insight.96560. JCI Insight. 2017. PMID: 29263295 Free PMC article.
-
Characterization of the chicken rhodopsin promoter: identification of retina-specific and glass-like protein binding domains.Mol Cell Neurosci. 1994 Aug;5(4):309-18. doi: 10.1006/mcne.1994.1037. Mol Cell Neurosci. 1994. PMID: 7804600
-
Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
-
Gene augmentation for adRP mutations in RHO.Cold Spring Harb Perspect Med. 2014 Jul 18;4(9):a017400. doi: 10.1101/cshperspect.a017400. Cold Spring Harb Perspect Med. 2014. PMID: 25037104 Free PMC article. Review.
Cited by
-
Progress in Gene Therapy for Rhodopsin Autosomal Dominant Retinitis Pigmentosa.Adv Exp Med Biol. 2019;1185:113-118. doi: 10.1007/978-3-030-27378-1_19. Adv Exp Med Biol. 2019. PMID: 31884598 Free PMC article. Review.
-
DNA residence time is a regulatory factor of transcription repression.Nucleic Acids Res. 2017 Nov 2;45(19):11121-11130. doi: 10.1093/nar/gkx728. Nucleic Acids Res. 2017. PMID: 28977492 Free PMC article.
-
Calpain Activation Is the Major Cause of Cell Death in Photoreceptors Expressing a Rhodopsin Misfolding Mutation.Mol Neurobiol. 2020 Feb;57(2):589-599. doi: 10.1007/s12035-019-01723-5. Epub 2019 Aug 10. Mol Neurobiol. 2020. PMID: 31401765
-
Revolution in Gene Medicine Therapy and Genome Surgery.Genes (Basel). 2018 Nov 26;9(12):575. doi: 10.3390/genes9120575. Genes (Basel). 2018. PMID: 30486314 Free PMC article. Review.
-
miR-181a/b downregulation exerts a protective action on mitochondrial disease models.EMBO Mol Med. 2019 May;11(5):e8734. doi: 10.15252/emmm.201708734. EMBO Mol Med. 2019. PMID: 30979712 Free PMC article.
References
-
- Daiger BR SP, Greenberg J, Christoffels A, Hide W. Data services and software for identifying genes and mutations causing retinal degeneration. Investigative Ophthalmology & Visual Science. 1998;39:S295.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

