Rab13 Traffics on Vesicles Independent of Prenylation
- PMID: 26969162
- PMCID: PMC4865919
- DOI: 10.1074/jbc.M116.722298
Rab13 Traffics on Vesicles Independent of Prenylation
Abstract
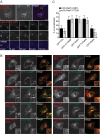
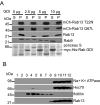
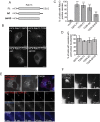
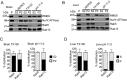
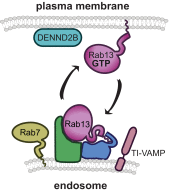
Rab GTPases are critical regulators of membrane trafficking. The canonical view is that Rabs are soluble in their inactive GDP-bound form, and only upon activation and conversion to their GTP-bound state are they anchored to membranes through membrane insertion of a C-terminal prenyl group. Here we demonstrate that C-terminal prenylation is not required for Rab13 to associate with and traffic on vesicles. Instead, inactive Rab13 appears to associate with vesicles via protein-protein interactions. Only following activation does Rab13 associate with the plasma membrane, presumably with insertion of the C-terminal prenyl group into the membrane.
Keywords: DENN domain; DENND2B; GDI; GDP dissociation inhibitor; Rab; TI-VAMP; endosome; guanine nucleotide exchange factor (GEF); protein isoprenylation; vesicles.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Yip3 catalyses the dissociation of endosomal Rab-GDI complexes.Nature. 2003 Oct 23;425(6960):856-9. doi: 10.1038/nature02057. Nature. 2003. PMID: 14574414
-
Molecular control of Rab activity by GEFs, GAPs and GDI.Small GTPases. 2018 Mar 4;9(1-2):5-21. doi: 10.1080/21541248.2016.1276999. Epub 2017 Feb 1. Small GTPases. 2018. PMID: 28055292 Free PMC article. Review.
-
The structural and mechanistic basis for recycling of Rab proteins between membrane compartments.Cell Mol Life Sci. 2005 Aug;62(15):1657-70. doi: 10.1007/s00018-005-4486-8. Cell Mol Life Sci. 2005. PMID: 15924270 Free PMC article. Review.
-
Membrane extraction of Rab proteins by GDP dissociation inhibitor characterized using attenuated total reflection infrared spectroscopy.Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13380-5. doi: 10.1073/pnas.1307655110. Epub 2013 Jul 29. Proc Natl Acad Sci U S A. 2013. PMID: 23898197 Free PMC article.
-
Disease mutations in Rab7 result in unregulated nucleotide exchange and inappropriate activation.Hum Mol Genet. 2010 Mar 15;19(6):1033-47. doi: 10.1093/hmg/ddp567. Epub 2009 Dec 22. Hum Mol Genet. 2010. PMID: 20028791 Free PMC article.
Cited by
-
Regulation of Cancer Cell Behavior by the Small GTPase Rab13.J Biol Chem. 2016 May 6;291(19):9929-37. doi: 10.1074/jbc.R116.715193. Epub 2016 Apr 4. J Biol Chem. 2016. PMID: 27044746 Free PMC article. Review.
-
Geranylgeranyl pyrophosphate depletion by statins compromises skeletal muscle insulin sensitivity.J Cachexia Sarcopenia Muscle. 2022 Dec;13(6):2697-2711. doi: 10.1002/jcsm.13061. Epub 2022 Aug 12. J Cachexia Sarcopenia Muscle. 2022. PMID: 35961942 Free PMC article.
-
Microtubules provide force to promote membrane uncoating in vacuolar escape for a cyto-invasive bacterial pathogen.Nat Commun. 2024 Feb 5;15(1):1065. doi: 10.1038/s41467-024-45182-6. Nat Commun. 2024. PMID: 38316786 Free PMC article.
-
Epstein-Barr virus subverts mevalonate and fatty acid pathways to promote infected B-cell proliferation and survival.PLoS Pathog. 2019 Sep 13;15(9):e1008030. doi: 10.1371/journal.ppat.1008030. eCollection 2019 Sep. PLoS Pathog. 2019. PMID: 31518366 Free PMC article.
-
Intersectin-s interaction with DENND2B facilitates recycling of epidermal growth factor receptor.EMBO Rep. 2017 Dec;18(12):2119-2130. doi: 10.15252/embr.201744034. Epub 2017 Oct 13. EMBO Rep. 2017. PMID: 29030480 Free PMC article.
References
-
- Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 - PubMed
-
- Yang P. S., Yin P. H., Tseng L. M., Yang C. H., Hsu C. Y., Lee M. Y., Horng C. F., and Chi C. W. (2011) Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 102, 2172–2178 - PubMed
-
- Allaire P. D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R. F., and McPherson P. S. (2013) Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126, 722–731 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

