Nucleosome repositioning underlies dynamic gene expression
- PMID: 26966245
- PMCID: PMC4803052
- DOI: 10.1101/gad.274910.115
Nucleosome repositioning underlies dynamic gene expression
Abstract
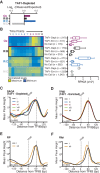
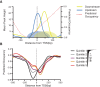
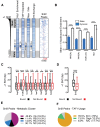
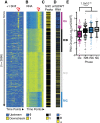
Nucleosome repositioning at gene promoters is a fundamental aspect of the regulation of gene expression. However, the extent to which nucleosome repositioning is used within eukaryotic genomes is poorly understood. Here we report a comprehensive analysis of nucleosome positions as budding yeast transit through an ultradian cycle in which expression of >50% of all genes is highly synchronized. We present evidence of extensive nucleosome repositioning at thousands of gene promoters as genes are activated and repressed. During activation, nucleosomes are relocated to allow sites of general transcription factor binding and transcription initiation to become accessible. The extent of nucleosome shifting is closely related to the dynamic range of gene transcription and generally related to DNA sequence properties and use of the coactivators TFIID or SAGA. However, dynamic gene expression is not limited to SAGA-regulated promoters and is an inherent feature of most genes. While nucleosome repositioning occurs pervasively, we found that a class of genes required for growth experience acute nucleosome shifting as cells enter the cell cycle. Significantly, our data identify that the ATP-dependent chromatin-remodeling enzyme Snf2 plays a fundamental role in nucleosome repositioning and the expression of growth genes. We also reveal that nucleosome organization changes extensively in concert with phases of the cell cycle, with large, regularly spaced nucleosome arrays being established in mitosis. Collectively, our data and analysis provide a framework for understanding nucleosome dynamics in relation to fundamental DNA-dependent transactions.
Keywords: SWI/SNF; chromatin; nucleosome; transcription.
© 2016 Nocetti and Whitehouse; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast.J Biol Chem. 2014 May 23;289(21):14981-95. doi: 10.1074/jbc.M113.529354. Epub 2014 Apr 11. J Biol Chem. 2014. PMID: 24727477 Free PMC article.
-
SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast.Genes Dev. 2018 May 1;32(9-10):695-710. doi: 10.1101/gad.312850.118. Epub 2018 May 21. Genes Dev. 2018. PMID: 29785963 Free PMC article.
-
Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters.Mol Biol Cell. 2011 Jun 15;22(12):2106-18. doi: 10.1091/mbc.E10-10-0826. Epub 2011 Apr 20. Mol Biol Cell. 2011. PMID: 21508315 Free PMC article.
-
Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence.Biochem Cell Biol. 2007 Aug;85(4):419-25. doi: 10.1139/O07-070. Biochem Cell Biol. 2007. PMID: 17713577 Review.
-
Primary Role of the Nucleosome.Mol Cell. 2020 Aug 6;79(3):371-375. doi: 10.1016/j.molcel.2020.07.020. Mol Cell. 2020. PMID: 32763226 Review.
Cited by
-
Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters.EMBO J. 2017 Feb 1;36(3):274-290. doi: 10.15252/embj.201695621. Epub 2016 Dec 15. EMBO J. 2017. PMID: 27979920 Free PMC article.
-
ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning.Genes (Basel). 2019 Sep 28;10(10):765. doi: 10.3390/genes10100765. Genes (Basel). 2019. PMID: 31569414 Free PMC article.
-
The nucleosome acidic patch directly interacts with subunits of the Paf1 and FACT complexes and controls chromatin architecture in vivo.Nucleic Acids Res. 2019 Sep 19;47(16):8410-8423. doi: 10.1093/nar/gkz549. Nucleic Acids Res. 2019. PMID: 31226204 Free PMC article.
-
The molecular basis of metabolic cycles and their relationship to circadian rhythms.Nat Struct Mol Biol. 2016 Dec 6;23(12):1035-1044. doi: 10.1038/nsmb.3311. Nat Struct Mol Biol. 2016. PMID: 27922609 Review.
-
An integrated machine-learning model to predict nucleosome architecture.Nucleic Acids Res. 2024 Sep 23;52(17):10132-10143. doi: 10.1093/nar/gkae689. Nucleic Acids Res. 2024. PMID: 39162225 Free PMC article.
References
-
- Cai L, Tu BP. 2012. Driving the cell cycle through metabolism. Annu Rev Cell Dev Biol 28: 59–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
