CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering
- PMID: 26953268
- PMCID: PMC4788126
- DOI: 10.1534/genetics.115.182162
CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering
Abstract
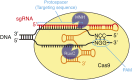
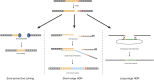
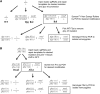
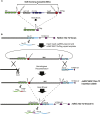
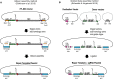
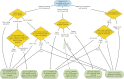
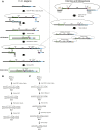
The advent of genome editing techniques based on the clustered regularly interspersed short palindromic repeats (CRISPR)-Cas9 system has revolutionized research in the biological sciences. CRISPR is quickly becoming an indispensible experimental tool for researchers using genetic model organisms, including the nematode Caenorhabditis elegans. Here, we provide an overview of CRISPR-based strategies for genome editing in C. elegans. We focus on practical considerations for successful genome editing, including a discussion of which strategies are best suited to producing different kinds of targeted genome modifications.
Keywords: CRISPR/Cas9; Caenorhabditis elegans; WormBook; genome editing.
Copyright © 2016 by the Genetics Society of America.
Figures
Similar articles
-
The application of somatic CRISPR-Cas9 to conditional genome editing in Caenorhabditis elegans.Genesis. 2016 Apr;54(4):170-81. doi: 10.1002/dvg.22932. Epub 2016 Apr 14. Genesis. 2016. PMID: 26934570 Review.
-
[CRISPR-Cas9 mediated genome editing in Caenorhabditis elegans].Sheng Wu Gong Cheng Xue Bao. 2017 Oct 25;33(10):1693-1699. doi: 10.13345/j.cjb.170177. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 29082717 Review. Chinese.
-
The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans.J Genet Genomics. 2015 Aug 20;42(8):413-21. doi: 10.1016/j.jgg.2015.06.005. Epub 2015 Jun 26. J Genet Genomics. 2015. PMID: 26336798 Free PMC article. Review.
-
Engineering the Caenorhabditis elegans genome with CRISPR/Cas9.Methods. 2014 Aug 1;68(3):381-8. doi: 10.1016/j.ymeth.2014.03.024. Epub 2014 Mar 28. Methods. 2014. PMID: 24685391
-
CRISPR-Cas9-Guided Genome Engineering in Caenorhabditis elegans.Curr Protoc Mol Biol. 2019 Dec;129(1):e106. doi: 10.1002/cpmb.106. Curr Protoc Mol Biol. 2019. PMID: 31763794 Free PMC article.
Cited by
-
Population scale nucleic acid delivery to Caenorhabditis elegans via electroporation.G3 (Bethesda). 2021 Jul 14;11(7):jkab123. doi: 10.1093/g3journal/jkab123. G3 (Bethesda). 2021. PMID: 33872353 Free PMC article.
-
FHOD-1 is the only formin in Caenorhabditis elegans that promotes striated muscle growth and Z-line organization in a cell autonomous manner.Cytoskeleton (Hoboken). 2020 Oct;77(10):422-441. doi: 10.1002/cm.21639. Epub 2020 Nov 6. Cytoskeleton (Hoboken). 2020. PMID: 33103378 Free PMC article.
-
Caenorhabditis elegans: a model to understand host-microbe interactions.Cell Mol Life Sci. 2020 Apr;77(7):1229-1249. doi: 10.1007/s00018-019-03319-7. Epub 2019 Oct 4. Cell Mol Life Sci. 2020. PMID: 31584128 Free PMC article. Review.
-
CRISPR interference for sequence-specific regulation of fibroblast growth factor receptor A in Schistosoma mansoni.Front Immunol. 2023 Jan 13;13:1105719. doi: 10.3389/fimmu.2022.1105719. eCollection 2022. Front Immunol. 2023. PMID: 36713455 Free PMC article.
-
A universal transportin protein drives stochastic choice of olfactory neurons via specific nuclear import of a sox-2-activating factor.Proc Natl Acad Sci U S A. 2019 Dec 10;116(50):25137-25146. doi: 10.1073/pnas.1908168116. Epub 2019 Nov 25. Proc Natl Acad Sci U S A. 2019. PMID: 31767767 Free PMC article.
References
-
- Carbone A., Zinovyev A., Képès F., 2003. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19: 2005–2015. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

