T cell fate and clonality inference from single-cell transcriptomes
- PMID: 26950746
- PMCID: PMC4835021
- DOI: 10.1038/nmeth.3800
T cell fate and clonality inference from single-cell transcriptomes
Abstract
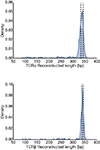
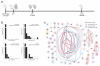
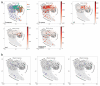
We developed TraCeR, a computational method to reconstruct full-length, paired T cell receptor (TCR) sequences from T lymphocyte single-cell RNA sequence data. TraCeR links T cell specificity with functional response by revealing clonal relationships between cells alongside their transcriptional profiles. We found that T cell clonotypes in a mouse Salmonella infection model span early activated CD4(+) T cells as well as mature effector and memory cells.
Figures
Similar articles
-
Antigen Receptor Sequence Reconstruction and Clonality Inference from scRNA-Seq Data.Methods Mol Biol. 2019;1935:223-249. doi: 10.1007/978-1-4939-9057-3_15. Methods Mol Biol. 2019. PMID: 30758830
-
Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis.Sci Immunol. 2018 Jan 19;3(19):eaan8664. doi: 10.1126/sciimmunol.aan8664. Sci Immunol. 2018. PMID: 29352091 Free PMC article.
-
Differences in TCR repertoire and T cell activation underlie the divergent outcomes of antitumor immune responses in tumor-eradicating versus tumor-progressing hosts.J Immunother Cancer. 2021 Jan;9(1):e001615. doi: 10.1136/jitc-2020-001615. J Immunother Cancer. 2021. PMID: 33414263 Free PMC article.
-
T cell receptor-transgenic mouse models for studying cellular immune responses to Salmonella in vivo.FEMS Immunol Med Microbiol. 2003 Jul 15;37(2-3):105-9. doi: 10.1016/S0928-8244(03)00064-6. FEMS Immunol Med Microbiol. 2003. PMID: 12832113 Review.
-
Revealing the heterogeneity of CD4+ T cells through single-cell transcriptomics.J Allergy Clin Immunol. 2022 Oct;150(4):748-755. doi: 10.1016/j.jaci.2022.08.010. J Allergy Clin Immunol. 2022. PMID: 36205449 Review.
Cited by
-
A Single-Cell Perspective on Memory T-Cell Differentiation.Cold Spring Harb Perspect Biol. 2021 Sep 1;13(9):a038067. doi: 10.1101/cshperspect.a038067. Cold Spring Harb Perspect Biol. 2021. PMID: 33903160 Free PMC article. Review.
-
Characterization and Monitoring of Antigen-Responsive T Cell Clones Using T Cell Receptor Gene Expression Analysis.Front Immunol. 2021 Feb 19;11:609624. doi: 10.3389/fimmu.2020.609624. eCollection 2020. Front Immunol. 2021. PMID: 33679697 Free PMC article.
-
Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery.Immunity. 2021 Mar 9;54(3):586-602.e8. doi: 10.1016/j.immuni.2021.02.014. Immunity. 2021. PMID: 33691136 Free PMC article.
-
Identification and Tracking of Alloreactive T Cell Clones in Rhesus Macaques Through the RM-scTCR-Seq Platform.Front Immunol. 2022 Jan 26;12:804932. doi: 10.3389/fimmu.2021.804932. eCollection 2021. Front Immunol. 2022. PMID: 35154078 Free PMC article.
-
Adaptive Immune Receptor Repertoire (AIRR) Community Guide to TR and IG Gene Annotation.Methods Mol Biol. 2022;2453:279-296. doi: 10.1007/978-1-0716-2115-8_16. Methods Mol Biol. 2022. PMID: 35622332 Free PMC article.
References
-
- Rossjohn J, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015;33:169–200. - PubMed
-
- Lieber MR. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. - PubMed
-
- Becattini S, et al. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. - PubMed
METHODS-ONLY REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

