Model-Driven Understanding of Palmitoylation Dynamics: Regulated Acylation of the Endoplasmic Reticulum Chaperone Calnexin
- PMID: 26900856
- PMCID: PMC4765739
- DOI: 10.1371/journal.pcbi.1004774
Model-Driven Understanding of Palmitoylation Dynamics: Regulated Acylation of the Endoplasmic Reticulum Chaperone Calnexin
Abstract
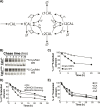
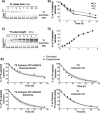
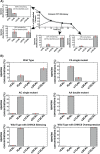
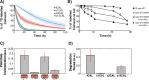
Cellular functions are largely regulated by reversible post-translational modifications of proteins which act as switches. Amongst these, S-palmitoylation is unique in that it confers hydrophobicity. Due to technical difficulties, the understanding of this modification has lagged behind. To investigate principles underlying dynamics and regulation of palmitoylation, we have here studied a key cellular protein, the ER chaperone calnexin, which requires dual palmitoylation for function. Apprehending the complex inter-conversion between single-, double- and non-palmitoylated species required combining experimental determination of kinetic parameters with extensive mathematical modelling. We found that calnexin, due to the presence of two cooperative sites, becomes stably acylated, which not only confers function but also a remarkable increase in stability. Unexpectedly, stochastic simulations revealed that palmitoylation does not occur soon after synthesis, but many hours later. This prediction guided us to find that phosphorylation actively delays calnexin palmitoylation in resting cells. Altogether this study reveals that cells synthesize 5 times more calnexin than needed under resting condition, most of which is degraded. This unused pool can be mobilized by preventing phosphorylation or increasing the activity of the palmitoyltransferase DHHC6.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Palmitoylated calnexin is a key component of the ribosome-translocon complex.EMBO J. 2012 Apr 4;31(7):1823-35. doi: 10.1038/emboj.2012.15. Epub 2012 Feb 7. EMBO J. 2012. PMID: 22314232 Free PMC article.
-
Palmitoylated TMX and calnexin target to the mitochondria-associated membrane.EMBO J. 2012 Jan 18;31(2):457-70. doi: 10.1038/emboj.2011.384. Epub 2011 Nov 1. EMBO J. 2012. PMID: 22045338 Free PMC article.
-
Palmitoylation is the switch that assigns calnexin to quality control or ER Ca2+ signaling.J Cell Sci. 2013 Sep 1;126(Pt 17):3893-903. doi: 10.1242/jcs.125856. Epub 2013 Jul 10. J Cell Sci. 2013. PMID: 23843619
-
Palmitoylation and palmitoyl-transferases in Plasmodium parasites.Biochem Soc Trans. 2015 Apr;43(2):240-5. doi: 10.1042/BST20140289. Biochem Soc Trans. 2015. PMID: 25849924 Review.
-
Selenoprotein K and protein palmitoylation.Antioxid Redox Signal. 2015 Oct 1;23(10):854-62. doi: 10.1089/ars.2015.6375. Epub 2015 Jun 17. Antioxid Redox Signal. 2015. PMID: 26058750 Free PMC article. Review.
Cited by
-
Protein Palmitoylation and Its Role in Bacterial and Viral Infections.Front Immunol. 2018 Jan 19;8:2003. doi: 10.3389/fimmu.2017.02003. eCollection 2017. Front Immunol. 2018. PMID: 29403483 Free PMC article. Review.
-
The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity.J Biol Chem. 2017 Dec 29;292(52):21517-21526. doi: 10.1074/jbc.M117.800482. Epub 2017 Oct 27. J Biol Chem. 2017. PMID: 29079573 Free PMC article.
-
Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade.Elife. 2017 Aug 15;6:e27826. doi: 10.7554/eLife.27826. Elife. 2017. PMID: 28826475 Free PMC article.
-
Endoplasmic Reticulum Stress in Metabolic Liver Diseases and Hepatic Fibrosis.Semin Liver Dis. 2019 May;39(2):235-248. doi: 10.1055/s-0039-1681032. Epub 2019 Mar 25. Semin Liver Dis. 2019. PMID: 30912096 Free PMC article. Review.
-
SARS-CoV-2 hijacks a cell damage response, which induces transcription of a more efficient Spike S-acyltransferase.Nat Commun. 2023 Nov 11;14(1):7302. doi: 10.1038/s41467-023-43027-2. Nat Commun. 2023. PMID: 37952051 Free PMC article.
References
-
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

