The UniProtKB guide to the human proteome
- PMID: 26896845
- PMCID: PMC4761109
- DOI: 10.1093/database/bav120
The UniProtKB guide to the human proteome
Abstract
Advances in high-throughput and advanced technologies allow researchers to routinely perform whole genome and proteome analysis. For this purpose, they need high-quality resources providing comprehensive gene and protein sets for their organisms of interest. Using the example of the human proteome, we will describe the content of a complete proteome in the UniProt Knowledgebase (UniProtKB). We will show how manual expert curation of UniProtKB/Swiss-Prot is complemented by expert-driven automatic annotation to build a comprehensive, high-quality and traceable resource. We will also illustrate how the complexity of the human proteome is captured and structured in UniProtKB. Database URL: www.uniprot.org.
© The Author(s) 2016. Published by Oxford University Press.
Figures

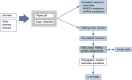
Similar articles
-
Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase.Hum Mutat. 2008 Mar;29(3):361-6. doi: 10.1002/humu.20671. Hum Mutat. 2008. PMID: 18175334
-
UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View.Methods Mol Biol. 2016;1374:23-54. doi: 10.1007/978-1-4939-3167-5_2. Methods Mol Biol. 2016. PMID: 26519399
-
The UniProtKB/Swiss-Prot knowledgebase and its Plant Proteome Annotation Program.J Proteomics. 2009 Apr 13;72(3):567-73. doi: 10.1016/j.jprot.2008.11.010. Epub 2008 Nov 24. J Proteomics. 2009. PMID: 19084081 Free PMC article.
-
Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation.Hum Mutat. 2014 Aug;35(8):927-35. doi: 10.1002/humu.22594. Epub 2014 Jun 24. Hum Mutat. 2014. PMID: 24848695 Free PMC article. Review.
-
Bioinformatics Tools for Proteomics Data Interpretation.Adv Exp Med Biol. 2016;919:281-341. doi: 10.1007/978-3-319-41448-5_16. Adv Exp Med Biol. 2016. PMID: 27975225 Review.
Cited by
-
Association Between aquaporin-1 and Endurance Performance: A Systematic Review.Sports Med Open. 2019 Sep 5;5(1):40. doi: 10.1186/s40798-019-0213-0. Sports Med Open. 2019. PMID: 31486928 Free PMC article. Review.
-
Functional dissection of human mitotic genes using CRISPR-Cas9 tiling screens.Genes Dev. 2022 Apr 1;36(7-8):495-510. doi: 10.1101/gad.349319.121. Epub 2022 Apr 28. Genes Dev. 2022. PMID: 35483740 Free PMC article.
-
Proteome changes of plasma-derived extracellular vesicles in patients with myelodysplastic syndrome.PLoS One. 2022 Jan 10;17(1):e0262484. doi: 10.1371/journal.pone.0262484. eCollection 2022. PLoS One. 2022. PMID: 35007303 Free PMC article.
-
Exploring the functional impact of alternative splicing on human protein isoforms using available annotation sources.Brief Bioinform. 2019 Sep 27;20(5):1754-1768. doi: 10.1093/bib/bby047. Brief Bioinform. 2019. PMID: 29931155 Free PMC article. Review.
-
Learning from low-rank multimodal representations for predicting disease-drug associations.BMC Med Inform Decis Mak. 2021 Nov 4;21(Suppl 1):308. doi: 10.1186/s12911-021-01648-x. BMC Med Inform Decis Mak. 2021. PMID: 34736437 Free PMC article.
References
-
- Wilhelm M., Schlegl J., Hahne H. et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature, 509, 582–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

