The crystal structure of human GlnRS provides basis for the development of neurological disorders
- PMID: 26869582
- PMCID: PMC4838373
- DOI: 10.1093/nar/gkw082
The crystal structure of human GlnRS provides basis for the development of neurological disorders
Abstract
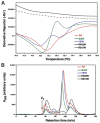
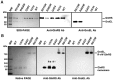
Cytosolic glutaminyl-tRNA synthetase (GlnRS) is the singular enzyme responsible for translation of glutamine codons. Compound heterozygous mutations in GlnRS cause severe brain disorders by a poorly understood mechanism. Herein, we present crystal structures of the wild type and two pathological mutants of human GlnRS, which reveal, for the first time, the domain organization of the intact enzyme and the structure of the functionally important N-terminal domain (NTD). Pathological mutations mapping in the NTD alter the domain structure, and decrease catalytic activity and stability of GlnRS, whereas missense mutations in the catalytic domain induce misfolding of the enzyme. Our results suggest that the reduced catalytic efficiency and a propensity of GlnRS mutants to misfold trigger the disease development. This report broadens the spectrum of brain pathologies elicited by protein misfolding and provides a paradigm for understanding the role of mutations in aminoacyl-tRNA synthetases in neurological diseases.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

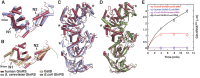


Similar articles
-
Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes.Int J Biol Macromol. 2018 Dec;120(Pt B):1379-1386. doi: 10.1016/j.ijbiomac.2018.09.115. Epub 2018 Sep 21. Int J Biol Macromol. 2018. PMID: 30248426
-
The structure of yeast glutaminyl-tRNA synthetase and modeling of its interaction with tRNA.J Mol Biol. 2013 Jul 24;425(14):2480-93. doi: 10.1016/j.jmb.2013.03.043. Epub 2013 Apr 10. J Mol Biol. 2013. PMID: 23583912 Free PMC article.
-
Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation.Nucleic Acids Res. 2007;35(5):1421-31. doi: 10.1093/nar/gkl1164. Epub 2007 Feb 6. Nucleic Acids Res. 2007. PMID: 17284460 Free PMC article.
-
Substrate selection by aminoacyl-tRNA synthetases.Nucleic Acids Symp Ser. 1995;(33):40-2. Nucleic Acids Symp Ser. 1995. PMID: 8643392 Review.
-
Glutaminyl-tRNA synthetase.Biol Chem. 1997 Oct;378(10):1103-17. Biol Chem. 1997. PMID: 9372179 Review.
Cited by
-
An optimized protocol for in vitro and in cellulo structural determination of the multi-tRNA synthetase complex by cross-linking mass spectrometry.STAR Protoc. 2022 Mar 9;3(1):101201. doi: 10.1016/j.xpro.2022.101201. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35284842 Free PMC article.
-
Influence of Disease-Causing Mutations on Protein Structural Networks.Front Mol Biosci. 2021 Mar 10;7:620554. doi: 10.3389/fmolb.2020.620554. eCollection 2020. Front Mol Biosci. 2021. PMID: 33778000 Free PMC article.
-
3-Dimensional architecture of the human multi-tRNA synthetase complex.Nucleic Acids Res. 2020 Sep 4;48(15):8740-8754. doi: 10.1093/nar/gkaa569. Nucleic Acids Res. 2020. PMID: 32644155 Free PMC article.
-
Defining and expanding the phenotype of QARS-associated developmental epileptic encephalopathy.Neurol Genet. 2019 Dec 10;5(6):e373. doi: 10.1212/NXG.0000000000000373. eCollection 2019 Dec. Neurol Genet. 2019. PMID: 32042906 Free PMC article.
-
Glutaminyl-tRNA Synthetase from Pseudomonas aeruginosa: Characterization, structure, and development as a screening platform.Protein Sci. 2020 Apr;29(4):905-918. doi: 10.1002/pro.3800. Epub 2019 Dec 24. Protein Sci. 2020. PMID: 31833153 Free PMC article.
References
-
- Ibba M., Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. - PubMed
-
- Schmidt E., Schimmel P. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science. 1994;264:265–267. - PubMed
-
- Mascarenhas A.P., An S., Rosen A.E., Martinis S.A., Musier-Forsyth K. In: Protein Engineering. RajBhandary UL, Köhrer C, editors. NY: Springer-Verlag; 2008. pp. 153–200.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

