Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan
- PMID: 26867182
- PMCID: PMC5106289
- DOI: 10.1016/j.chom.2016.01.008
Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan
Abstract
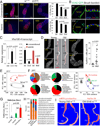
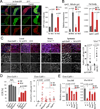
Compartmentalization of the gastrointestinal (GI) tract of metazoans is critical for health. GI compartments contain specific microbiota, and microbiota dysbiosis is associated with intestinal dysfunction. Dysbiosis develops in aging intestines, yet how this relates to changes in GI compartmentalization remains unclear. The Drosophila GI tract is an accessible model to address this question. Here we show that the stomach-like copper cell region (CCR) in the middle midgut controls distribution and composition of the microbiota. We find that chronic activation of JAK/Stat signaling in the aging gut induces a metaplasia of the gastric epithelium, CCR decline, and subsequent commensal dysbiosis and epithelial dysplasia along the GI tract. Accordingly, inhibition of JAK/Stat signaling in the CCR specifically prevents age-related metaplasia, commensal dysbiosis and functional decline in old guts, and extends lifespan. Our results establish a mechanism by which age-related chronic inflammation causes the decline of intestinal compartmentalization and microbiota dysbiosis, limiting lifespan.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Microbes without Borders: Decompartmentalization of the Aging Gut.Cell Host Microbe. 2016 Feb 10;19(2):133-5. doi: 10.1016/j.chom.2016.01.016. Cell Host Microbe. 2016. PMID: 26867169
Similar articles
-
Vitamin D Receptor Protects Against Dysbiosis and Tumorigenesis via the JAK/STAT Pathway in Intestine.Cell Mol Gastroenterol Hepatol. 2020;10(4):729-746. doi: 10.1016/j.jcmgh.2020.05.010. Epub 2020 Jun 1. Cell Mol Gastroenterol Hepatol. 2020. PMID: 32497792 Free PMC article.
-
Tools and methods for studying the Drosophila JAK/STAT pathway.Methods. 2014 Jun 15;68(1):160-72. doi: 10.1016/j.ymeth.2014.03.023. Epub 2014 Mar 28. Methods. 2014. PMID: 24685392 Review.
-
High-fat-diet-induced modulations of leptin signaling and gastric microbiota drive precancerous lesions in the stomach.Nutrition. 2019 Nov-Dec;67-68:110556. doi: 10.1016/j.nut.2019.110556. Epub 2019 Jul 23. Nutrition. 2019. PMID: 31554603
-
Eubiosis and dysbiosis: the two sides of the microbiota.New Microbiol. 2016 Jan;39(1):1-12. New Microbiol. 2016. PMID: 26922981 Review.
-
Dysbiosis of gut microbiota and microbial metabolites in Parkinson's Disease.Ageing Res Rev. 2018 Aug;45:53-61. doi: 10.1016/j.arr.2018.04.004. Epub 2018 Apr 26. Ageing Res Rev. 2018. PMID: 29705121 Review.
Cited by
-
Cellular senescence: Implications for metabolic disease.Mol Cell Endocrinol. 2017 Nov 5;455:93-102. doi: 10.1016/j.mce.2016.08.047. Epub 2016 Aug 31. Mol Cell Endocrinol. 2017. PMID: 27591120 Free PMC article. Review.
-
The Intestine of Drosophila melanogaster: An Emerging Versatile Model System to Study Intestinal Epithelial Homeostasis and Host-Microbial Interactions in Humans.Microorganisms. 2019 Sep 9;7(9):336. doi: 10.3390/microorganisms7090336. Microorganisms. 2019. PMID: 31505811 Free PMC article. Review.
-
Intestinal probiotics restore the ecological fitness decline of Bactrocera dorsalis by irradiation.Evol Appl. 2018 Oct 9;11(10):1946-1963. doi: 10.1111/eva.12698. eCollection 2018 Dec. Evol Appl. 2018. PMID: 30459840 Free PMC article.
-
Model systems for regeneration: Drosophila.Development. 2020 Apr 6;147(7):dev173781. doi: 10.1242/dev.173781. Development. 2020. PMID: 32253254 Free PMC article. Review.
-
How does a fly die? Insights into ageing from the pathophysiology of Drosophila mortality.Geroscience. 2024 Oct;46(5):4003-4015. doi: 10.1007/s11357-024-01158-4. Epub 2024 Apr 20. Geroscience. 2024. PMID: 38642259 Free PMC article. Review.
References
-
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene expression patterns : GEP. 2007;7:323–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

