Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo
- PMID: 26858442
- PMCID: PMC4763755
- DOI: 10.1073/pnas.1522675113
Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo
Abstract
Reservoirs of infectious HIV-1 persist despite years of combination antiretroviral therapy and make curing HIV-1 infections a major challenge. Most of the proviral DNA resides in CD4(+)T cells. Some of these CD4(+)T cells are clonally expanded; most of the proviruses are defective. It is not known if any of the clonally expanded cells carry replication-competent proviruses. We report that a highly expanded CD4(+) T-cell clone contains an intact provirus. The highly expanded clone produced infectious virus that was detected as persistent plasma viremia during cART in an HIV-1-infected patient who had squamous cell cancer. Cells containing the intact provirus were widely distributed and significantly enriched in cancer metastases. These results show that clonally expanded CD4(+)T cells can be a reservoir of infectious HIV-1.
Keywords: HIV persistence; clonal expansion of infected cells; replication-competent HIV.
Conflict of interest statement
Conflict of interest statement: J.W.M. is a consultant for Gilead Sciences and owns shares in Co-Crystal, Inc.
Figures

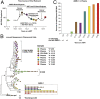
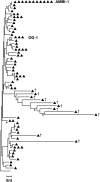
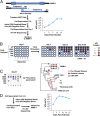
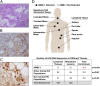
Comment in
-
Reservoir expansion by T-cell proliferation may be another barrier to curing HIV infection.Proc Natl Acad Sci U S A. 2016 Feb 16;113(7):1692-4. doi: 10.1073/pnas.1600097113. Epub 2016 Feb 9. Proc Natl Acad Sci U S A. 2016. PMID: 26862166 Free PMC article. No abstract available.
Similar articles
-
CD161+ CD4+ T Cells Harbor Clonally Expanded Replication-Competent HIV-1 in Antiretroviral Therapy-Suppressed Individuals.mBio. 2019 Oct 8;10(5):e02121-19. doi: 10.1128/mBio.02121-19. mBio. 2019. PMID: 31594817 Free PMC article.
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
-
Intact HIV Proviruses Persist in Children Seven to Nine Years after Initiation of Antiretroviral Therapy in the First Year of Life.J Virol. 2020 Jan 31;94(4):e01519-19. doi: 10.1128/JVI.01519-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776265 Free PMC article.
-
Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections.Viruses. 2020 Jan 25;12(2):136. doi: 10.3390/v12020136. Viruses. 2020. PMID: 31991737 Free PMC article.
-
New Approaches to Multi-Parametric HIV-1 Genetics Using Multiple Displacement Amplification: Determining the What, How, and Where of the HIV-1 Reservoir.Viruses. 2021 Dec 10;13(12):2475. doi: 10.3390/v13122475. Viruses. 2021. PMID: 34960744 Free PMC article. Review.
Cited by
-
HIV infected CD4+ T cell clones are more stable than uninfected clones during long-term antiretroviral therapy.PLoS Pathog. 2022 Aug 31;18(8):e1010726. doi: 10.1371/journal.ppat.1010726. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 36044447 Free PMC article.
-
HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo.Cells. 2022 Sep 23;11(19):2968. doi: 10.3390/cells11192968. Cells. 2022. PMID: 36230931 Free PMC article.
-
The Latent Reservoir for HIV-1: How Immunologic Memory and Clonal Expansion Contribute to HIV-1 Persistence.J Immunol. 2016 Jul 15;197(2):407-17. doi: 10.4049/jimmunol.1600343. J Immunol. 2016. PMID: 27382129 Free PMC article. Review.
-
Transcription of HIV-1 at sites of intact latent provirus integration.J Exp Med. 2024 Sep 2;221(9):e20240391. doi: 10.1084/jem.20240391. Epub 2024 Aug 14. J Exp Med. 2024. PMID: 39141127 Free PMC article.
-
Assessing the impact of autologous virus neutralizing antibodies on viral rebound time in postnatally SHIV-infected ART-treated infant rhesus macaques.Epidemics. 2024 Sep;48:100780. doi: 10.1016/j.epidem.2024.100780. Epub 2024 Jun 27. Epidemics. 2024. PMID: 38964130 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

