Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx
- PMID: 26858413
- PMCID: PMC4776483
- DOI: 10.1073/pnas.1525616113
Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx
Abstract
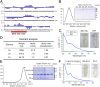
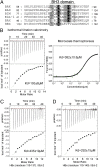
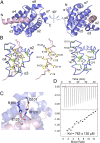
HBx is a hepatitis B virus protein that is required for viral infectivity and replication. Anti-apoptotic Bcl-2 family members are thought to be among the important host targets of HBx. However, the structure and function of HBx are poorly understood and the molecular mechanism of HBx-induced carcinogenesis remains unknown. In this study, we report biochemical and structural characterization of HBx. The recombinant HBx protein contains metal ions, in particular iron and zinc. A BH3-like motif in HBx (residues 110-135) binds Bcl-2 with a dissociation constant of ∼193 μM, which is drastically lower than that for a canonical BH3 motif from Bim or Bad. Structural analysis reveals that, similar to other BH3 motifs, the BH3-like motif of HBx adopts an amphipathic α-helix and binds the conserved BH3-binding groove on Bcl-2. Unlike the helical Bim or Bad BH3 motif, the C-terminal portion of the bound HBx BH3-like motif has an extended conformation and makes considerably fewer interactions with Bcl-2. These observations suggest that HBx may modulate Bcl-2 function in a way that is different from that of the classical BH3-only proteins.
Keywords: Bcl-2; apoptosis; crystal structure; hepatitis B virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structural and functional analyses of hepatitis B virus X protein BH3-like domain and Bcl-xL interaction.Nat Commun. 2019 Jul 19;10(1):3192. doi: 10.1038/s41467-019-11173-1. Nat Commun. 2019. PMID: 31324803 Free PMC article.
-
Structural characterization of the BH3-like motif of hepatitis B virus X protein.Biochem Biophys Res Commun. 2014 Jul 18;450(1):741-5. doi: 10.1016/j.bbrc.2014.06.042. Epub 2014 Jun 17. Biochem Biophys Res Commun. 2014. PMID: 24950407
-
NMR characterization of the interaction between Bcl-xL and the BH3-like motif of hepatitis B virus X protein.Biochem Biophys Res Commun. 2019 Oct 20;518(3):445-450. doi: 10.1016/j.bbrc.2019.08.036. Epub 2019 Aug 19. Biochem Biophys Res Commun. 2019. PMID: 31439373
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
-
Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma.J Gastroenterol Hepatol. 2011 Jan;26 Suppl 1:144-52. doi: 10.1111/j.1440-1746.2010.06546.x. J Gastroenterol Hepatol. 2011. PMID: 21199526 Review.
Cited by
-
Computational Evaluation of Abrogation of HBx-Bcl-xL Complex with High-Affinity Carbon Nanotubes (Fullerene) to Halt the Hepatitis B Virus Replication.Molecules. 2021 Oct 25;26(21):6433. doi: 10.3390/molecules26216433. Molecules. 2021. PMID: 34770842 Free PMC article.
-
Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6.Viruses. 2017 Apr 3;9(4):69. doi: 10.3390/v9040069. Viruses. 2017. PMID: 28368357 Free PMC article. Review.
-
Hepatitis B Virus X Protein Function Requires Zinc Binding.J Virol. 2019 Jul 30;93(16):e00250-19. doi: 10.1128/JVI.00250-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31167910 Free PMC article.
-
Thyroid hormone protects hepatocytes from HBx-induced carcinogenesis by enhancing mitochondrial turnover.Oncogene. 2017 Sep 14;36(37):5274-5284. doi: 10.1038/onc.2017.136. Epub 2017 May 15. Oncogene. 2017. PMID: 28504722
-
Pathogenicity and virulence of Hepatitis B virus.Virulence. 2022 Dec;13(1):258-296. doi: 10.1080/21505594.2022.2028483. Virulence. 2022. PMID: 35100095 Free PMC article. Review.
References
-
- Benhenda S, Cougot D, Buendia MA, Neuveut C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res. 2009;103:75–109. - PubMed
-
- Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: Paradigms for viral-related human carcinogenesis. Oncogene. 2006;25(27):3823–3833. - PubMed
-
- Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34(Suppl 1):S75–S78. - PubMed
-
- Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

