MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production
- PMID: 26850657
- PMCID: PMC4838571
- DOI: 10.1016/j.immuni.2016.01.003
MicroRNAs 24 and 27 Suppress Allergic Inflammation and Target a Network of Regulators of T Helper 2 Cell-Associated Cytokine Production
Abstract
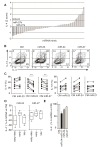
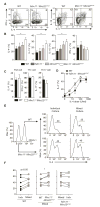
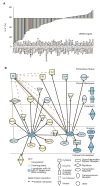
MicroRNAs (miRNAs) are important regulators of cell fate decisions in immune responses. They act by coordinate repression of multiple target genes, a property that we exploited to uncover regulatory networks that govern T helper-2 (Th2) cells. A functional screen of individual miRNAs in primary T cells uncovered multiple miRNAs that inhibited Th2 cell differentiation. Among these were miR-24 and miR-27, miRNAs coexpressed from two genomic clusters, which each functioned independently to limit interleukin-4 (IL-4) production. Mice lacking both clusters in T cells displayed increased Th2 cell responses and tissue pathology in a mouse model of asthma. Gene expression and pathway analyses placed miR-27 upstream of genes known to regulate Th2 cells. They also identified targets not previously associated with Th2 cell biology which regulated IL-4 production in unbiased functional testing. Thus, elucidating the biological function and target repertoire of miR-24 and miR-27 reveals regulators of Th2 cell biology.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



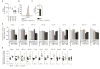
Similar articles
-
miR-23∼27∼24 clusters control effector T cell differentiation and function.J Exp Med. 2016 Feb 8;213(2):235-49. doi: 10.1084/jem.20150990. Epub 2016 Feb 1. J Exp Med. 2016. PMID: 26834155 Free PMC article.
-
MicroRNA-155 modulates P2R signaling and Th2 priming of dendritic cells during allergic airway inflammation in mice.Allergy. 2015 Sep;70(9):1121-9. doi: 10.1111/all.12643. Epub 2015 May 28. Allergy. 2015. PMID: 25944053
-
A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production.Nat Immunol. 2014 Dec;15(12):1162-70. doi: 10.1038/ni.3026. Epub 2014 Nov 2. Nat Immunol. 2014. PMID: 25362490 Free PMC article.
-
T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production.Cytokine. 2015 Sep;75(1):14-24. doi: 10.1016/j.cyto.2015.05.010. Epub 2015 Jun 1. Cytokine. 2015. PMID: 26044597 Free PMC article. Review.
-
The impact of microRNAs on alterations of gene regulatory networks in allergic diseases.Adv Protein Chem Struct Biol. 2020;120:237-312. doi: 10.1016/bs.apcsb.2019.11.006. Epub 2020 Feb 12. Adv Protein Chem Struct Biol. 2020. PMID: 32085883 Review.
Cited by
-
MicroRNAs in blood and cerebrospinal fluid as diagnostic biomarkers of multiple sclerosis and to monitor disease progression.Neural Regen Res. 2020 Apr;15(4):606-619. doi: 10.4103/1673-5374.266905. Neural Regen Res. 2020. PMID: 31638082 Free PMC article. Review.
-
An essential role for miR-15/16 in Treg suppression and restriction of proliferation.Cell Rep. 2023 Oct 31;42(10):113298. doi: 10.1016/j.celrep.2023.113298. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37862171 Free PMC article.
-
MicroRNA-directed pathway discovery elucidates an miR-221/222-mediated regulatory circuit in class switch recombination.J Exp Med. 2021 Nov 1;218(11):e20201422. doi: 10.1084/jem.20201422. Epub 2021 Sep 29. J Exp Med. 2021. PMID: 34586363 Free PMC article.
-
Non-coding RNA Contribution to Thoracic and Abdominal Aortic Aneurysm Disease Development and Progression.Front Physiol. 2017 Jun 16;8:429. doi: 10.3389/fphys.2017.00429. eCollection 2017. Front Physiol. 2017. PMID: 28670289 Free PMC article. Review.
-
Sevoflurane induces inflammation in primary hippocampal neurons by regulating Hoxa5/Gm5106/miR-27b-3p positive feedback loop.Bioengineered. 2021 Dec;12(2):12215-12226. doi: 10.1080/21655979.2021.2005927. Bioengineered. 2021. PMID: 34783294 Free PMC article.
References
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

