RNA Structure Duplications and Flavivirus Host Adaptation
- PMID: 26850219
- PMCID: PMC4808370
- DOI: 10.1016/j.tim.2016.01.002
RNA Structure Duplications and Flavivirus Host Adaptation
Abstract
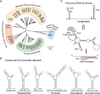
Flaviviruses include a highly diverse group of arboviruses with a global distribution and a high human disease burden. Most flaviviruses cycle between insects and vertebrate hosts; thus, they are obligated to use different cellular machinery for their replication and mount different mechanisms to evade specific antiviral responses. In addition to coding for viral proteins, the viral genome contains signals in RNA structures that govern the amplification of viral components and participate in triggering or evading antiviral responses. In this review, we focused on new information about host-specific functions of RNA structures present in the 3' untranslated region (3' UTR) of flavivirus genomes. Models and conservation patterns of RNA elements of distinct flavivirus ecological groups are revised. An intriguing feature of the 3' UTR of insect-borne flavivirus genomes is the conservation of complex RNA structure duplications. Here, we discuss new hypotheses of how these RNA elements specialize for replication in vertebrate and invertebrate hosts, and present new ideas associating the significance of RNA structure duplication, small subgenomic flavivirus RNA formation, and host adaptation.
Keywords: RNA virus evolution; dengue virus; flaviviruses; host adaptation; sfRNAs; viral RNA structures.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures





Similar articles
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation.RNA Biol. 2015;12(11):1169-77. doi: 10.1080/15476286.2015.1094599. Epub 2015 Sep 23. RNA Biol. 2015. PMID: 26399159 Free PMC article. Review.
-
RNA Structure Duplication in the Dengue Virus 3' UTR: Redundancy or Host Specificity?mBio. 2019 Jan 8;10(1):e02506-18. doi: 10.1128/mBio.02506-18. mBio. 2019. PMID: 30622191 Free PMC article.
-
Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells.PLoS Pathog. 2017 Mar 6;13(3):e1006265. doi: 10.1371/journal.ppat.1006265. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28264033 Free PMC article.
-
Subgenomic flaviviral RNAs: What do we know after the first decade of research.Antiviral Res. 2018 Nov;159:13-25. doi: 10.1016/j.antiviral.2018.09.006. Epub 2018 Sep 11. Antiviral Res. 2018. PMID: 30217649 Review.
Cited by
-
Zika Virus Pathogenesis: A Battle for Immune Evasion.Vaccines (Basel). 2021 Mar 22;9(3):294. doi: 10.3390/vaccines9030294. Vaccines (Basel). 2021. PMID: 33810028 Free PMC article. Review.
-
Short Direct Repeats in the 3' Untranslated Region Are Involved in Subgenomic Flaviviral RNA Production.J Virol. 2020 Feb 28;94(6):e01175-19. doi: 10.1128/JVI.01175-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896596 Free PMC article.
-
Dengue and Zika Virus 5' Untranslated Regions Harbor Internal Ribosomal Entry Site Functions.mBio. 2019 Apr 9;10(2):e00459-19. doi: 10.1128/mBio.00459-19. mBio. 2019. PMID: 30967466 Free PMC article.
-
Full-Genome Characterization and Genetic Evolution of West African Isolates of Bagaza Virus.Viruses. 2018 Apr 13;10(4):193. doi: 10.3390/v10040193. Viruses. 2018. PMID: 29652824 Free PMC article.
-
Role of RNA-binding proteins during the late stages of Flavivirus replication cycle.Virol J. 2020 Apr 25;17(1):60. doi: 10.1186/s12985-020-01329-7. Virol J. 2020. PMID: 32334603 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

