Chromosome-scale shotgun assembly using an in vitro method for long-range linkage
- PMID: 26848124
- PMCID: PMC4772016
- DOI: 10.1101/gr.193474.115
Chromosome-scale shotgun assembly using an in vitro method for long-range linkage
Abstract
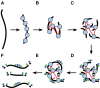
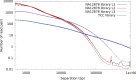
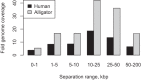
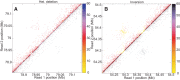
Long-range and highly accurate de novo assembly from short-read data is one of the most pressing challenges in genomics. Recently, it has been shown that read pairs generated by proximity ligation of DNA in chromatin of living tissue can address this problem, dramatically increasing the scaffold contiguity of assemblies. Here, we describe a simpler approach ("Chicago") based on in vitro reconstituted chromatin. We generated two Chicago data sets with human DNA and developed a statistical model and a new software pipeline ("HiRise") that can identify poor quality joins and produce accurate, long-range sequence scaffolds. We used these to construct a highly accurate de novo assembly and scaffolding of a human genome with scaffold N50 of 20 Mbp. We also demonstrated the utility of Chicago for improving existing assemblies by reassembling and scaffolding the genome of the American alligator. With a single library and one lane of Illumina HiSeq sequencing, we increased the scaffold N50 of the American alligator from 508 kbp to 10 Mbp.
© 2016 Putnam et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Similar articles
-
In vitro, long-range sequence information for de novo genome assembly via transposase contiguity.Genome Res. 2014 Dec;24(12):2041-9. doi: 10.1101/gr.178319.114. Epub 2014 Oct 19. Genome Res. 2014. PMID: 25327137 Free PMC article.
-
Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary.Mol Ecol Resour. 2019 Jul;19(4):1015-1026. doi: 10.1111/1755-0998.13020. Epub 2019 May 17. Mol Ecol Resour. 2019. PMID: 30972949 Free PMC article.
-
An integrated chromosome-scale genome assembly of the Masai giraffe (Giraffa camelopardalis tippelskirchi).Gigascience. 2019 Aug 1;8(8):giz090. doi: 10.1093/gigascience/giz090. Gigascience. 2019. PMID: 31367745 Free PMC article.
-
New Approaches for Genome Assembly and Scaffolding.Annu Rev Anim Biosci. 2019 Feb 15;7:17-40. doi: 10.1146/annurev-animal-020518-115344. Epub 2018 Nov 28. Annu Rev Anim Biosci. 2019. PMID: 30485757 Review.
-
Personalized genome structure via single gamete sequencing.Genome Biol. 2021 Apr 19;22(1):112. doi: 10.1186/s13059-021-02327-w. Genome Biol. 2021. PMID: 33874978 Free PMC article. Review.
Cited by
-
Draft Genomes of Amaranthus tuberculatus, Amaranthus hybridus, and Amaranthus palmeri.Genome Biol Evol. 2020 Nov 3;12(11):1988-1993. doi: 10.1093/gbe/evaa177. Genome Biol Evol. 2020. PMID: 32835372 Free PMC article.
-
Construction of Pseudomolecules for the Chinese Chestnut (Castanea mollissima) Genome.G3 (Bethesda). 2020 Oct 5;10(10):3565-3574. doi: 10.1534/g3.120.401532. G3 (Bethesda). 2020. PMID: 32847817 Free PMC article.
-
Chromosome-scale genome assembly of the brown anole (Anolis sagrei), an emerging model species.Commun Biol. 2022 Oct 25;5(1):1126. doi: 10.1038/s42003-022-04074-5. Commun Biol. 2022. PMID: 36284162 Free PMC article.
-
Sequencing of Camelina neglecta, a diploid progenitor of the hexaploid oilseed Camelina sativa.Plant Biotechnol J. 2023 Mar;21(3):521-535. doi: 10.1111/pbi.13968. Epub 2022 Dec 12. Plant Biotechnol J. 2023. PMID: 36398722 Free PMC article.
-
A high-quality, haplotype-phased genome reconstruction reveals unexpected haplotype diversity in a pearl oyster.DNA Res. 2022 Dec 1;29(6):dsac035. doi: 10.1093/dnares/dsac035. DNA Res. 2022. PMID: 36351462 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
