Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies
- PMID: 26831084
- PMCID: PMC4763790
- DOI: 10.1073/pnas.1519906113
Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies
Abstract
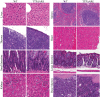
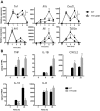
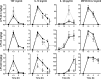
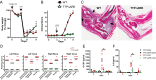
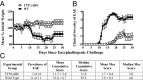
Tristetraprolin (TTP) is an inducible, tandem zinc-finger mRNA binding protein that binds to adenylate-uridylate-rich elements (AREs) in the 3'-untranslated regions (3'UTRs) of specific mRNAs, such as that encoding TNF, and increases their rates of deadenylation and turnover. Stabilization of Tnf mRNA and other cytokine transcripts in TTP-deficient mice results in the development of a profound, chronic inflammatory syndrome characterized by polyarticular arthritis, dermatitis, myeloid hyperplasia, and autoimmunity. To address the hypothesis that increasing endogenous levels of TTP in an intact animal might be beneficial in the treatment of inflammatory diseases, we generated a mouse model (TTPΔARE) in which a 136-base instability motif in the 3'UTR of TTP mRNA was deleted in the endogenous genetic locus. These mice appeared normal, but cultured fibroblasts and macrophages derived from them exhibited increased stability of the otherwise highly labile TTP mRNA. This resulted in increased TTP protein expression in LPS-stimulated macrophages and increased levels of TTP protein in mouse tissues. TTPΔARE mice were protected from collagen antibody-induced arthritis, exhibited significantly reduced inflammation in imiquimod-induced dermatitis, and were resistant to induction of experimental autoimmune encephalomyelitis, presumably by dampening the excessive production of proinflammatory mediators in all cases. These data suggest that increased systemic levels of TTP, secondary to increased stability of its mRNA throughout the body, can be protective against inflammatory disease in certain models and might be viewed as an attractive therapeutic target for the treatment of human inflammatory diseases.
Keywords: AU-rich elements; deadenylation; inflammation; mRNA stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
3'UTR AU-Rich Elements (AREs) and the RNA-Binding Protein Tristetraprolin (TTP) Are Not Required for the LPS-Mediated Destabilization of Phospholipase-Cβ-2 mRNA in Murine Macrophages.Inflammation. 2017 Apr;40(2):645-656. doi: 10.1007/s10753-017-0511-y. Inflammation. 2017. PMID: 28124257 Free PMC article.
-
Tristetraprolin overexpression drives hematopoietic changes in young and middle-aged mice generating dominant mitigating effects on induced inflammation in murine models.Geroscience. 2024 Feb;46(1):1271-1284. doi: 10.1007/s11357-023-00879-2. Epub 2023 Aug 3. Geroscience. 2024. PMID: 37535204 Free PMC article.
-
Regulated Tristetraprolin Overexpression Dampens the Development and Pathogenesis of Experimental Autoimmune Uveitis.Front Immunol. 2021 Jan 25;11:583510. doi: 10.3389/fimmu.2020.583510. eCollection 2020. Front Immunol. 2021. PMID: 33569048 Free PMC article.
-
Clinical implications of tristetraprolin (TTP) modulation in the treatment of inflammatory diseases.Pharmacol Ther. 2022 Nov;239:108198. doi: 10.1016/j.pharmthera.2022.108198. Epub 2022 May 5. Pharmacol Ther. 2022. PMID: 35525391 Free PMC article. Review.
-
Tristetraprolin as a Therapeutic Target in Inflammatory Disease.Trends Pharmacol Sci. 2016 Oct;37(10):811-821. doi: 10.1016/j.tips.2016.07.002. Epub 2016 Aug 5. Trends Pharmacol Sci. 2016. PMID: 27503556 Free PMC article. Review.
Cited by
-
Tristetraprolin limits age-related expansion of myeloid-derived suppressor cells.Front Immunol. 2022 Oct 3;13:1002163. doi: 10.3389/fimmu.2022.1002163. eCollection 2022. Front Immunol. 2022. PMID: 36263047 Free PMC article.
-
Microglial Nox2 Plays a Key Role in the Pathogenesis of Experimental Autoimmune Encephalomyelitis.Front Immunol. 2021 Apr 2;12:638381. doi: 10.3389/fimmu.2021.638381. eCollection 2021. Front Immunol. 2021. PMID: 33868265 Free PMC article.
-
RNA-Binding Protein ZFP36L2 Downregulates Helios Expression and Suppresses the Function of Regulatory T Cells.Front Immunol. 2020 Jun 23;11:1291. doi: 10.3389/fimmu.2020.01291. eCollection 2020. Front Immunol. 2020. PMID: 32655569 Free PMC article.
-
Uncovering the Role of RNA-Binding Proteins in Gene Expression in the Immune System.Front Immunol. 2018 May 23;9:1094. doi: 10.3389/fimmu.2018.01094. eCollection 2018. Front Immunol. 2018. PMID: 29875770 Free PMC article. Review.
-
Improving Dietary Intake of Essential Nutrients Can Ameliorate Inflammation in Patients with Diabetic Foot Ulcers.Nutrients. 2022 Jun 9;14(12):2393. doi: 10.3390/nu14122393. Nutrients. 2022. PMID: 35745123 Free PMC article. Clinical Trial.
References
-
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30(Pt 6):945–952. - PubMed
-
- Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281(5379):1001–1005. - PubMed
-
- Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98(8):2389–2395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

