A new view of transcriptome complexity and regulation through the lens of local splicing variations
- PMID: 26829591
- PMCID: PMC4801060
- DOI: 10.7554/eLife.11752
A new view of transcriptome complexity and regulation through the lens of local splicing variations
Abstract
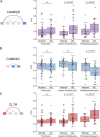
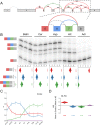
Alternative splicing (AS) can critically affect gene function and disease, yet mapping splicing variations remains a challenge. Here, we propose a new approach to define and quantify mRNA splicing in units of local splicing variations (LSVs). LSVs capture previously defined types of alternative splicing as well as more complex transcript variations. Building the first genome wide map of LSVs from twelve mouse tissues, we find complex LSVs constitute over 30% of tissue dependent transcript variations and affect specific protein families. We show the prevalence of complex LSVs is conserved in humans and identify hundreds of LSVs that are specific to brain subregions or altered in Alzheimer's patients. Amongst those are novel isoforms in the Camk2 family and a novel poison exon in Ptbp1, a key splice factor in neurogenesis. We anticipate the approach presented here will advance the ability to relate tissue-specific splice variation to genetic variation, phenotype, and disease.
Keywords: alternative splicing; alzheimer's disease; camk2; chromosomes; computational biology; genes; human; mouse; ptb1; statistical modeling; systems biology; transcriptomics.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










Similar articles
-
Discover hidden splicing variations by mapping personal transcriptomes to personal genomes.Nucleic Acids Res. 2015 Dec 15;43(22):10612-22. doi: 10.1093/nar/gkv1099. Epub 2015 Nov 17. Nucleic Acids Res. 2015. PMID: 26578562 Free PMC article.
-
Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer's disease susceptibility.Nat Genet. 2018 Nov;50(11):1584-1592. doi: 10.1038/s41588-018-0238-1. Epub 2018 Oct 8. Nat Genet. 2018. PMID: 30297968 Free PMC article.
-
Mapping Whole-Transcriptome Splicing in Mouse Hematopoietic Stem Cells.Stem Cell Reports. 2017 Jan 10;8(1):163-176. doi: 10.1016/j.stemcr.2016.12.002. Epub 2016 Dec 29. Stem Cell Reports. 2017. PMID: 28041879 Free PMC article.
-
Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases.Neurobiol Aging. 2012 May;33(5):1012.e11-24. doi: 10.1016/j.neurobiolaging.2011.10.030. Epub 2011 Nov 26. Neurobiol Aging. 2012. PMID: 22118946 Review.
-
Alternative splicing: the pledge, the turn, and the prestige : The key role of alternative splicing in human biological systems.Hum Genet. 2017 Sep;136(9):1015-1042. doi: 10.1007/s00439-017-1790-y. Epub 2017 Apr 3. Hum Genet. 2017. PMID: 28374191 Free PMC article. Review.
Cited by
-
Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus.BMC Biol. 2023 Nov 9;21(1):251. doi: 10.1186/s12915-023-01756-2. BMC Biol. 2023. PMID: 37946231 Free PMC article.
-
Serine/arginine-rich splicing factor 7 promotes the type I interferon response by activating Irf7 transcription.Cell Rep. 2024 Mar 26;43(3):113816. doi: 10.1016/j.celrep.2024.113816. Epub 2024 Feb 22. Cell Rep. 2024. PMID: 38393946 Free PMC article.
-
SpliceWiz: interactive analysis and visualization of alternative splicing in R.Brief Bioinform. 2023 Nov 22;25(1):bbad468. doi: 10.1093/bib/bbad468. Brief Bioinform. 2023. PMID: 38152981 Free PMC article.
-
Alternative splicing of HDAC7 regulates its interaction with 14-3-3 proteins to alter histone marks and target gene expression.Cell Rep. 2023 Mar 28;42(3):112273. doi: 10.1016/j.celrep.2023.112273. Epub 2023 Mar 17. Cell Rep. 2023. PMID: 36933216 Free PMC article.
-
TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A.Nature. 2022 Mar;603(7899):131-137. doi: 10.1038/s41586-022-04436-3. Epub 2022 Feb 23. Nature. 2022. PMID: 35197628 Free PMC article.
References
-
- Bai B, Hales CM, Chen P-C, Gozal Y, Dammer EB, Fritz JJ, Wang X, Xia Q, Duong DM, Street C, Cantero G, Cheng D, Jones DR, Wu Z, Li Y, Diner I, Heilman CJ, Rees HD, Wu H, Lin L, Szulwach KE, Gearing M, Mufson EJ, Bennett DA, Montine TJ, Seyfried NT, Wingo TS, Sun YE, Jin P, Hanfelt J, Willcock DM, Levey A, Lah JJ, Peng J, Xia Q, Duong DM, Street C. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16562–16567. doi: 10.1073/pnas.1310249110. - DOI - PMC - PubMed
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

